Abstract
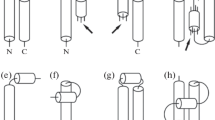
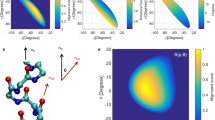
This paper presents the results and discussion of statistical analysis of helix-helix interactions in proteins in terms of interhelical angle and sequential distance. The protein data includes 1290 high-resolution globular protein structures with less than 20 percent homology. We first define possible cases of interactions between helices and identify protein pairs that satisfy the conditions for each case. Two cases of helix-helix interactions are investigated using six parameters that can be obtained by considering the interactions as rigid-body motions. The computational results show that orientational preferences in helix packing are dominated by interactions between helices that are proximal in sequence. This bias is described with a univariate probability density function that depends only on the sequential distance between interacting helices. We also compute bivariate probability densities that depend on both interhelical orientation and sequential distance. The dominance of certain preferred orientations and sequential distance values in this bivariate distribution are also observed. Angle preferences persist even when the bivariate distribution is normalized by the univariate distribution. This statistical analysis provides quantitative probability distributions from which statistical potentials for secondary structure assembly can be derived.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Baldwin, R.L., Rose, G.D.: Is protein folding hierarchic? I. Local structure and peptide folding. Trends in Biochemical Sciences 24(1), 26–33 (1999)
Baldwin, R.L., Rose, G.D.: Is protein folding hierarchic? II. Folding intermediates and transition states. Trends in Biochemical Sciences 24(2), 77–83 (1999)
Crick, F.: The packing of α-helical: simple coiled coils. Acta Crystallogr 6, 689–697 (1953)
Chothia, C., Levitt, M., Richardson, D.: Structure of proteins: Packing of (-helices and pleated sheets. Proc. Natl. Acad. Sci. USA 74(10), 4130–4134 (1977)
Chothia, C., Levitt, M., Richardson, D.: Helix to helix packing in proteins. J. Mol. Biol. 145, 215–250 (1981)
Finkelstein, A.V., Ptitsyn, O.B.: Why do globular proteins fit the limited set of folding patterns? Prog. Biophys. Molec. Biol. 50, 171–190 (1987)
Murzin, A.G., Finkelstein, A.V.: General architecture of the α-helical: globule. J. Mol. Biol. 204, 749–769 (1988)
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., et al.: The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000)
Bowie, J.U.: Helix packing angle preferences. Nature Structural Biology 4(11), 915–917 (1997)
Walther, D., Springer, C., Cohen, F.E.: Helix-helix packing angle preferences for finite helix axes. Proteins: Structure, Function and Genetics 33, 457–459 (1998)
Lee, S., Chirikjian, G.S.: Interhelical Angle and Distance Preferences in Globular Proteins. Biophysical Journal 86, 1105–1117 (2004)
Wang, G., Dunbrack, R.L.: PISCES: a Protein Sequence Culling Server. Bioinformatics 19, 1589–1591 (2003)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2008 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Lee, S. (2008). Analysis of Interhelical Angle and Sequential Distance in Proteins. In: Gervasi, O., Murgante, B., Laganà, A., Taniar, D., Mun, Y., Gavrilova, M.L. (eds) Computational Science and Its Applications – ICCSA 2008. ICCSA 2008. Lecture Notes in Computer Science, vol 5073. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-69848-7_86
Download citation
DOI: https://doi.org/10.1007/978-3-540-69848-7_86
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-69840-1
Online ISBN: 978-3-540-69848-7
eBook Packages: Computer ScienceComputer Science (R0)