Abstract
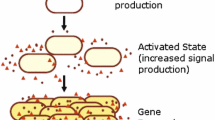
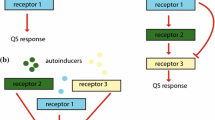
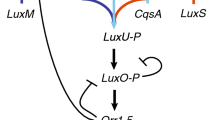
“Quorum sensing” has been described as “the most consequential molecular microbiology story of the last decade” [1][2]. The purpose of this paper is to study the mechanism of quorum sensing, in order to obtain a deeper understanding of how and when this mechanism works. Our study focuses on the use of an Individual-based Modeling (IbM) method to simulate this phenomenon of “cell-to-cell communication” incorporated in bacterial foraging behavior, in both intracellular and population scales. The simulation results show that this IbM approach can reflect the bacterial behaviors and population evolution in time-varying environments, and provide plausible answers to the emerging question regarding to the significance of this phenomenon of bacterial foraging behaviors.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Busby, S., de Lorenzo, V.: Cell regulation putting together pieces of the big puzzle. Current Opinion in Microbiology 5, 117–118 (2001)
Winzer, K., Hardie, K.R., Williams, P.: Bacterial cell-to-cell communication: sorry, can’t talk now - gone to lunch! Current Opinion in Microbiology 5, 216–222 (2002)
Vlachos, C., Paton, R.C., Saunders, J.R., Wu, Q.H.: A rule-based approach to the modelling of bacterial ecosystems. BioSystems 84, 49–72 (2005)
Ben-Jacob, E., Shochet, O., Tenenbaum, A., Cohen, I.: Generic modeling of cooperative growth patterns in bacterial colonies. Nature 368, 46–49 (1994)
Davies, D.G., Parsek, M.R., Pearson, J.P. et al.: The involvement of cell-to-cell signals in the development of a bacterial biofilm. Nature 280, 295–298 (1998)
Ward, J.P., King, J.R., Koerber, A.J.: Mathematical modelling of quorum sensing in bacteria. Journal of Mathematics Applied in Medicine and Biology 18, 263–292 (2001)
Dockery, J.D., Keener, J.P.: A mathematical model for quorum sensing in pseudomonas aeruginosa. Mathematical Biology, 1–22 (2000)
Painter, K.J., Hillen, T.: Volume-filling and quorum sensing in models for chemosensitive movement. Canadian Applied Mathematics Quarterly 10, 51–543 (2002)
Goryachev, A.B., Toh, D.J., Wee, K.B., et al.: Transition to quorum sensing in an agrobacterium population: A stochastic model. PLoS Computational Biology, 1–51 (2005)
Muller, J., Kuttler, C., Hense, B.A.: Cell-cell communication by quorum sensing and dimension-reduction. Technical Report, Technical University Munich, pp. 1–28 (2005)
You, L., Cox III, R.C., Weiss, R., Arnold, F. H.: Programmed population control by cell-cell communication and regulated killing. Nature (2004)
Garcia-Ojalvo, J., Elowitz, M.B., Strogatz, S.H.: Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing. Proceedings of the National Academy of Sciences 101, 10955–10960 (2004)
Tang, W.J., Wu, Q.H., Saunders, J.R.: A novel model for bacterial foraging in varying environments. In: Gavrilova, M.L., Gervasi, O., Kumar, V., Tan, C.J.K., Taniar, D., Laganá, A., Mun, Y., Choo, H. (eds.) ICCSA 2006. LNCS, vol. 3980, pp. 556–565. Springer, Heidelberg (2006)
Miller, M.B., Bassler, B.L.: Quorum sensing in bacteria. Annual Review of Microbiology 55, 165–199 (2001)
Ester, M., Kriegel, H., Sander, J., Xu, X.: A density-based algorithm for discovering clusters in large spatical databases with noise. In: Proceedings of 2nd International Conference on Knowledge Discovery and Data Mining, KDD-96 (1996)
Cao, Y.J., Wu, Q.H.: Study of initial population in evolutionary programming. In: Proceedings of the European Control Conference, vol. 368, pp. 1–4 (1997)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2007 Springer Berlin Heidelberg
About this paper
Cite this paper
Tang, W.J., Wu, Q.H., Saunders, J.R. (2007). Individual-Based Modeling of Bacterial Foraging with Quorum Sensing in a Time-Varying Environment. In: Marchiori, E., Moore, J.H., Rajapakse, J.C. (eds) Evolutionary Computation,Machine Learning and Data Mining in Bioinformatics. EvoBIO 2007. Lecture Notes in Computer Science, vol 4447. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-71783-6_27
Download citation
DOI: https://doi.org/10.1007/978-3-540-71783-6_27
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-71782-9
Online ISBN: 978-3-540-71783-6
eBook Packages: Computer ScienceComputer Science (R0)