Abstract
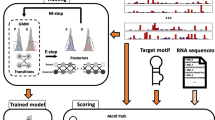
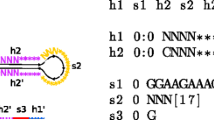
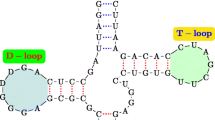
In this paper we present the design and implementation of an RNA structural motif database, called RmotifDB. The structural motifs stored in RmotifDB come from three sources: (1) collected manually from biomedical literature; (2) submitted by scientists around the world; and (3) discovered by a wide variety of motif mining methods. We present here a motif mining method in detail. We also describe the interface and search mechanisms provided by RmotifDB as well as techniques used to integrate RmotifDB with the Gene Ontology. The RmotifDB system is fully operational and accessible on the Internet at http://datalab.njit.edu/bioinfo/
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Akmaev, V.R., Kelley, S.T., Stormo, G.D.: Phylogenetically enhanced statistical tools for RNA structure prediction. Bioinformatics 16, 501–512 (2000)
Bakheet, T., Frevel, M., Williams, B.R., Greer, W., Khabar, K.S.: ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29, 246–254 (2001)
Bindewald, E., Shapiro, B.A.: RNA secondary structure prediction from sequence alignments using a network of k-nearest neighbor classifiers. RNA 12, 342–352 (2006)
Bindewald, E., Schneider, T.D., Shapiro, B.A.: CorreLogo: an online server for 3D sequence logos of RNA and DNA alignments. Nucleic Acids Res. 34, 405–411 (2006)
Cohen.-Boulakia, S., Davidson, S.B., Froidevaux, C.: A user-centric framework for accessing biological sources and tools. In: Proc. of the 2nd International Workshop on Data Integration in the Life Sciences, pp. 3–18 (2005)
Dalgaard, P.: Introductory Statistics with R. Springer, Heidelberg (2004)
Davidson, S.B., Crabtree, J., Brunk, B.P., Schug, J., Tannen, V., Overton, G.C., Stoeckert Jr., C.J.: K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Systems Journal 40, 512–531 (2001)
Eddy, S.R.: A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 3(18) (2002)
Gorodkin, J., Stricklin, S.L., Stormo, G.D.: Discovering common stem-loop motifs in unaligned RNA sequences. Nucleic Acids Res. 29, 2135–2144 (2001)
Grillo, G., Licciulli, F., Liuni, S., Sbisa, E., Pesole, G.: PatSearch: a program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 31, 3608–3612 (2003)
Hofacker, I.L.: Vienna RNA secondary structure server. Nucleic Acids Res. 31, 3429–3431 (2003)
Jones, S.G., Moxon, S., Marshall, M., Khanna, A., Eddy, S.R., Bateman, A.: Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33,D121–D124 (2005)
Khaladkar, M., Bellofatto, V., Wang, J.T.L., Tian, B., Zhang, K.: RADAR: an interactive web-based toolkit for RNA data analysis and research. In: Proc. of the 6th IEEE Symposium on Bioinformatics and Bioengineering, pp. 209–212. IEEE Computer Society Press, Los Alamitos (2006)
Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B.: Prediction of mammalian microRNA targets. Cell. 115, 787–798 (2003)
Liu, J., Wang, J.T.L., Hu, J., Tian, B.: A method for aligning RNA secondary structures and its application to RNA motif detection. BMC Bioinformatics 6(89) (2005)
Mignone, F., Grillo, G., Licciulli, F., Iacono, M., Liuni, S., Kersey, P.J., Duarte, J., Saccone, C., Pesole, G.: UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 33, 141–146 (2005)
Pruitt, K.D., Katz, K.S., Sicotte, H., Maglott, D.R.: Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet 16, 44–47 (2000)
Wang, J.T.L., Rozen, S., Shapiro, B.A., Shasha, D., Wang, Z., Yin, M.: New techniques for DNA sequence classification. Journal of Computational Biology 6, 209–218 (1999)
Wang, J.T.L., Shapiro, B.A., Shasha, D., Zhang, K., Currey, K.M.: An algorithm for finding the largest approximately common substructures of two trees. IEEE Transactions on Pattern Analysis and Machine Intelligence 20, 889–895 (1998)
Wang, J.T.L., Wu, X.: Kernel design for RNA classification using support vector machines. International Journal of Data Mining and Bioinformatics 1, 57–76 (2006)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2007 Springer Berlin Heidelberg
About this paper
Cite this paper
Wang, J.T.L., Wen, D., Shapiro, B.A., Herbert, K.G., Li, J., Ghosh, K. (2007). Toward an Integrated RNA Motif Database. In: Cohen-Boulakia, S., Tannen, V. (eds) Data Integration in the Life Sciences. DILS 2007. Lecture Notes in Computer Science(), vol 4544. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-73255-6_5
Download citation
DOI: https://doi.org/10.1007/978-3-540-73255-6_5
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-73254-9
Online ISBN: 978-3-540-73255-6
eBook Packages: Computer ScienceComputer Science (R0)