Abstract
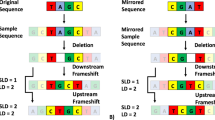
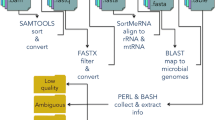
In order to improve EST trimming, we proposed a new method consisting of a new set of procedures to detect regions that do not belong to the sequenced organism or have low quality or low complexity. Most trimming procedures process ESTs in a pipeline where the output of an step is adopted as the input for the following one. In our method, all artifact detection steps process the raw EST and their results are combined in the last step, which outputs the trimmed sequence. This strategy reduces the occurrence of false negatives and, additionally, has the advantage of producing better artifact composition characterization for the analyzed sequences. We evaluated our method using SUCEST [1] ESTs. Based on the results, we concluded that our method suits projects that want to produce more reliable clusters.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Vettore, A.L., da Silva, F.R., Kemper, E.L., Arruda, P.: The libraries that made SUCEST. Genetics and Molecular Biology 406, 151–157 (2001)
Adams, M.D., Kelley, J.M., Gocayne, J.D., Dubnick, M., Polymeropoulos, M.H., Xiao, H., Merril, C.R., Wu, A., Olde, B., Moreno, R.F., Kerlavage, A.R., McCombie, W.R., Venter, J.C.: Complementary DNA Sequencing: Expressed Sequence Tags and Human Genome Project. Science 252, 1651–1656 (1991)
Telles, G.P., da Silva, F.R.: Trimming and clustering sugarcane ESTs. Genetics and Molecular Biology 24(1-4), 17–23 (2001)
Scheetz, T.E., Trivedi, N., Roberts, C.A., Kucaba, T., Berger, B., Robinson, N.L., Birkett, C.L., Gavin, A.J., O’Leary, B., Braun, T.A., Bonaldo, M.F., Robinson, H.P., Sheffield, V.C., Soares, M.B., Casavant, T.L.: ESTprep: preprocessing cDNA sequence. Bioinformatics 19(11), 1318–1324 (2003)
Chou, H., Holmes, M.H.: DNA sequence quality trimming and vector removal. Bioinformatics 17, 1093–1104 (2001)
Baudet, C., Dias, Z.: New EST Trimming Strategy. In: Setubal, J.C., Verjovski-Almeida, S. (eds.) BSB 2005. LNCS (LNBI), vol. 3594, pp. 206–209. Springer, Heidelberg (2005)
Band, M.R., Larson, J.H., Rebeiz, M., Green, C.A., Heyen, D.W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J.E., Lewin, H.A.: An Ordered Comparative Map of the Cattle and Human Genomes. Genome Research 10, 1359–1368 (2000)
Cattle EST Project: The W. M. Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign (January 2005), http://titan.biotec.uiuc.edu/cattle/cattle_project.htm
Ma, R.Z., van Eijk, M.J.T., Beever, J.E., Guérin, G., Mummery, C.L., Lewin, H.A.: Comparative analysis of 82 expressed sequence tags from a cattle ovary cDNA library. Mammalian Genome 9, 545–549 (1998)
Baudet, C., Dias, Z.: Analysis of slipped sequences in EST projects. Genetics and Molecular Research 5(1), 169–181 (2006)
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: Basic local alignment search tool. Journal of Molecular Biology 215(3), 403–410 (1990)
Manber, U.: Introduction to Algorithms. Addison-Wesley, Reading (1989)
Ewing, B., Hillier, L., Wendl, M.C., Green, P.: Base-Calling of Automated Sequencer Traces Using Phred. I. Accuracy Assessment. Genome Research 8(3), 175–185 (1998)
Ewing, B., Green, P.: Base-Calling of Automated Sequencer Traces Using Phred. II. Error Probabilities. Genome Research 8(3), 186–194 (1998)
Green, P.: Phrap Homepage: phred, phrap, consed, swat, cross_match and Repeat-Masker Documentation (March 2004), http://www.phrap.org
Huang, X., Madan, A.: CAP3: a DNA sequence assembly program. Genome Research 9, 868–877 (1999)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2007 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Baudet, C., Dias, Z. (2007). New EST Trimming Procedure Applied to SUCEST Sequences. In: Sagot, MF., Walter, M.E.M.T. (eds) Advances in Bioinformatics and Computational Biology. BSB 2007. Lecture Notes in Computer Science(), vol 4643. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-73731-5_6
Download citation
DOI: https://doi.org/10.1007/978-3-540-73731-5_6
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-73730-8
Online ISBN: 978-3-540-73731-5
eBook Packages: Computer ScienceComputer Science (R0)