Abstract
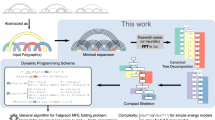
Predicting the secondary structure of an RNA sequence is an important problem in structural bioinformatics. The general RNA folding problem, where the sequence to be folded may contain pseudoknots, is computationally intractable when no prior knowledge on the pseudoknot structures the sequence contains is available. In this paper, we consider stable stems in an RNA sequence and provide a new characterization for its stem graph, a graph theoretic model that has been used to describe the overlapping relationships for stable stems. Based on this characterization, we identify a new structure parameter for a stem graph. We call this structure parameter crossing width. We show that given a sequence with crossing width c for its stem graph, the general RNA folding problem can be solved in time O(2c k 3 n 2), where n is the length of the sequence, k is the maximum length of stable stems. Moreover, this characterization leads to an \(O(2^{(1+2k^2)n}n^{2}k^{3})\) time algorithm for the general RNA folding problem where the lengths of stems in the sequence are at most k, this result improves the upper bound of the problem to 2O(n) n 2 when the maximum stem length is bounded by a constant.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Abrahams, J., van den Berg, M., van Batenburg, E., Pleij, C.: Prediction of RNA secondary structure including pseudoknotting, by computer simulation. Nucleic Acids Research 18, 3035–3044 (1990)
Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., Strobel, S.A.: Crystal structure of a self-splicing group i intron with both exons. Nature 430, 4550 (2004)
Akutsu, T.: Dynamic programming algorithms for RNA secondary structure prediction with pseudoknots. Discrete Applied Mathematics 104, 45–62 (2000)
Bodlaender, H.L.: Dynamic programming algorithms on graphs with bounded tree-width. In: Lepistö, T., Salomaa, A. (eds.) Automata, Languages and Programming. LNCS, vol. 317, pp. 105–119. Springer, Heidelberg (1988)
Cai, L., Malmberg, R.L., Wu, Y.: Stochastic modeling of pseudoknotted structures: a grammatical approach. In: Proceedings of the 11th International Conference on Intelligent Systems for Molecular Biology, pp. 66–73 (2003)
Chen, J.-H., Le, S.-Y., Maize, J.V.: Prediction of common secondary structures of RNAs: a genetic algorithm approach. Nucleic Acids Research 28(4), 991–999 (2000)
Gilmore, P.C., Hoffman, A.J.: A characterization of comparability graphs and of interval graphs. Canadian Journal of Mathematics 16(99), 539–548 (1964)
Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S.R., Bateman, A.: Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Research 33, D121–D124 (2005)
Ke, A., Zhou, K., Ding, F., Gate, J.H., Doudna, J.A.: A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature 429, 201–205 (2004)
Lyngso, R.B., Pedersen, C.N.S.: RNA pseudoknot prediction in energy-based models. Journal of Computational Biology 7(3-4), 409–427 (2000)
Available at: http://www.bioinfo.rpi.edu/zukerm/rna/energy/node2.html#SECTION20
Nussinov, R., Pieczenik, G., Griggs, J., Kleitman, D.: Algorithms for loop matchings. SIAM Journal of Applied Mathematics 35, 68–82 (1978)
Reeder, J., Giegerich, R.: Design, Implementation and Evaluation of A Practical Pseudoknot Folding Algorithm Based on Thermodynamics. BMC Bioinformatics 5, 104 (2004)
Ren, J., Rastegart, B., Condon, A., Hoos, H.H.: HotKnots: Heuristic prediction of RNA structures including pseudoknots. RNA 11, 1194–1504 (2005)
Rivas, E., Eddy, S.R.: A dynamic programming algorithm for RNA structure prediction including pseudoknots. Journal of Molecular Biology 285, 2053–2068 (1999)
Robertson, N., Seymour, P.D.: Graph Minors II: Algorithmic aspects of tree width. Journal of Algorithms 7, 309–322 (1986)
Ruan, J., Stormo, G.D., Zhang, W.: An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics 20(1), 58–66 (2004)
Schimmel, P.: RNA pseudoknots that interact with components of the translation apparatus. Cell 58(1), 9–12 (1989)
Tabaska, J., Gary, R., Gabow, H., Stormo, G.: An RNA folding method capable of identifying pseudoknots and base triples. Bioinformatics 14(8), 691–699 (1998)
Uemura, Y., Hasegawa, A., Kobayashi, S., Yokomori, T.: Tree adjoining grammars for RNA structure prediction. Theoretical Computer Science 210, 277–303 (1999)
Zuker, M., Stiegler, P.: Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Research 9(1), 133–148 (1981)
Zhao, J., Malmberg, R.L., Cai, L.: Rapid ab initio RNA folding including pseudoknots via graph tree decomposition. In: Bücher, P., Moret, B.M.E. (eds.) WABI 2006. LNCS (LNBI), vol. 4175, pp. 262–273. Springer, Heidelberg (2006)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2007 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Liu, C., Song, Y., Shapiro, L. (2007). RNA Folding Including Pseudoknots: A New Parameterized Algorithm and Improved Upper Bound. In: Giancarlo, R., Hannenhalli, S. (eds) Algorithms in Bioinformatics. WABI 2007. Lecture Notes in Computer Science(), vol 4645. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-74126-8_29
Download citation
DOI: https://doi.org/10.1007/978-3-540-74126-8_29
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-74125-1
Online ISBN: 978-3-540-74126-8
eBook Packages: Computer ScienceComputer Science (R0)