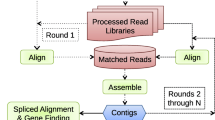
Abstract
Next Generation Sequencing (NGS) technologies are capable of reading millions of short DNA sequences both quickly and cheaply. While these technologies are already being used for resequencing individuals once a reference genome exists, it has not been shown if it is possible to use them for ab initio genome assembly. In this paper, we give a novel network flow-based algorithm that, by taking advantage of the high coverage provided by NGS, accurately estimates the copy counts of repeats in a genome. We also give a second algorithm that combines the predicted copy-counts with mate-pair data in order to assemble the reads into contigs. We run our algorithms on simulated read data from E. Coli and predict copy-counts with extremely high accuracy, while assembling long contigs.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Ahuja, R.K., Magnanti, T.L., Orlin, J.B.: Network flows: theory, algorithms, and applications. Prentice-Hall, Inc., Upper Saddle River, NJ, USA (1993)
Appa, G., Kotnyek, B.: A bidirected generalization of network matrices. Networks 47(4), 185–198 (2006)
Batzoglou, S., Jaffe, D.B., Stanley, K., Butler, J., Gnerre, S., Mauceli, E., Berger, B., Mesirov, J.P., Lander, E.S.: Arachne: a whole-genome shotgun assembler. Genome Res. 12(1), 177–189 (2002)
Chaisson, M., Pevzner, P.A., Tang, H.: Fragment assembly with short reads. Bioinformatics 20(13), 2067–2074 (2004)
Chaisson, M.J., Pevzner, P.A.: Short read fragment assembly of bacterial genomes. Genome. Published online before print Doi:10.1101/gr.7088808 (2007)
Dohm, J., Lottaz, C., Borodina, T., Himmelbauer, H.: SHARCGS, a fast and highly accurate short-read assembly algorithm for de novo genomic sequencing. Genome Res. 17, 1697–1706 (2007)
Edmonds, J.: An introduction to matching. In: Notes of engineering summer conference, University of Michigan, Ann Arbor (1967)
Gabow, H.N.: An efficient reduction technique for degree-constrained subgraph and bidirected network flow problems. In: STOC, pp. 448–456 (1983)
Goldberg, A.V.: An efficient implementation of a scaling minimum-cost flow algorithm. J. Algorithms 22(1), 1–29 (1997)
Hochbaum, D.S.: Monotonizing linear programs with up to two nonzeroes per column. Oper. Res. Lett. 32(1), 49–58 (2004)
Chen, J., Skiena, S.: Assembly For Double-Ended Short-Read Sequencing Technologies. In: Mardis, E., Kim, S., Tang, H. (eds.) Advances in Genome Sequencing Technology and Algorithms, pp. 123–141. Artech House Publishers (2007)
Jeck, W.R., Reinhardt, J.A., Baltrus, D.A., Hickenbotham, M.T., Magrini, V., Mardis, E.R., Dangl, J.L., Jones, C.D.: Extending assembly of short DNA sequences to handle error. Bioinformatics 23, 2942–2944 (2007)
Kececioglu, J.D.: Exact and approximation algorithms for DNA sequence reconstruction. PhD thesis, University of Arizona, Tucson, AZ, USA (1992)
Medvedev, P., Georgiou, K., Myers, G., Brudno, M.: Computability of models for sequence assembly. In: WABI, pp. 289–301 (2007)
Myers, E.W.: The fragment assembly string graph. In: ECCB/JBI, p. 85 (2005)
Myers, E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H.J., Remington, K.A., Anson, E.L., Bolanos, R.A., Chou, H.-H., Jordan, C.M., Halpern, A.L., Lonardi, S., Beasley, E.M., Brandon, R.C., Chen, L., Dunn, P.J., Lai, Z., Liang, Y., Nusskern, D.R., Zhan, M., Zhang, Q., Zheng, X., Rubin, G.M., Adams, M.D., Venter, J.C.: A Whole-Genome Assembly of Drosophila. Science 287, 2196–2204 (2000)
Pevzner, P.A., Tang, H., Waterman, M.S.: An Eulerian path approach to DNA fragment assembly. Proceedings of the National Academy of Sciences 98, 9748–9753 (2001)
Pevzner, P.A., Tang, H.: Fragment assembly with double-barreled data. In: ISMB (Supplement of Bioinformatics), pp. 225–233 (2001)
Pevzner, P.A., Tang, H., Tesler, G.: De novo repeat classification and fragment assembly. In: RECOMB, pp. 213–222 (2004)
Warren, R.L., Sutton, G.G., Jones, S.J., Holt, R.A.: Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23, 500–501 (2007)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2008 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Medvedev, P., Brudno, M. (2008). Ab Initio Whole Genome Shotgun Assembly with Mated Short Reads. In: Vingron, M., Wong, L. (eds) Research in Computational Molecular Biology. RECOMB 2008. Lecture Notes in Computer Science(), vol 4955. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-78839-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-540-78839-3_5
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-78838-6
Online ISBN: 978-3-540-78839-3
eBook Packages: Computer ScienceComputer Science (R0)