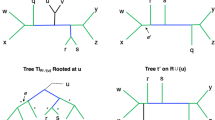
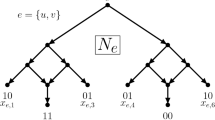
Abstract
The Multi-State Perfect Phylogeny Problem is an extension of the Binary Perfect Phylogeny Problem, allowing characters to take on more than two states. In this paper we consider three problems that extend the utility of the multi-state perfect phylogeny model: The Missing Data (MD) Problem where some entries in the input are missing and the question is whether (bounded) values for the missing data can be imputed so that the resulting data has a multi-state perfect phylogeny; The Character-Removal (CR) Problem where we want to minimize the number of characters to remove from the data so that the resulting data has a multi-state perfect phylogeny; and The Missing-Data Character-Removal (MDCR) Problem where the input has missing data and we want to impute values for the missing data to minimize the solution to the resulting Character-Removal Problem.
We detail Integer Linear Programming (ILP) solutions to these problems for the special case of three permitted states per character and report on extensive empirical testing of these solutions. Then we develop a general theory to solve the MD problem for an arbitrary number of permitted states, using chordal graph theory and results on minimal triangulation of non-chordal graphs. This establishes new necessary and sufficient conditions for the existence of a perfect phylogeny with (or without) missing data. We implement the general theory using integer linear programming, although other optimization methods are possible. We extensively explore the empirical behavior of the general solution, showing that the methods are very practical for data of size and complexity that is characteristic of many current applications in phylogenetics. Some of the empirical results for the MD problem with an arbitrary number of permitted states are very surprising, suggesting the existence of additional combinatorial structure in multi-state perfect phylogenies.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Agarwala, R., Fernandez-Baca, D.: A polynomial-time algorithm for the perfect phylogeny problem when the number of character states is fixed. SIAM J. on Computing 23, 1216–1224 (1994)
Berry, A., Bordat, J.-P., Cogis, O.: Generating All the Minimal Separators of a Graph. In: Widmayer, P., Neyer, G., Eidenbenz, S. (eds.) WG 1999. LNCS, vol. 1665, pp. 167–172. Springer, Heidelberg (1999)
Bodlaender, H., Fellows, M., Warnow, T.: Two strikes against perfect phylogeny. In: Proc. of the 19’th Inter. colloquium on Automata, Languages and Programming, pp. 273–283 (1992)
Buneman, P.: A characterization of rigid circuit graphs. Discrete Math. 9, 205–212 (1974)
Dress, A., Steel, M.: Convex tree realizations of partitions. Applied Math. Letters 5, 3–6 (1993)
Estabrook, G., Johnson, C., McMorris, F.: An idealized concept of the true cladistic character. Math. Bioscience 23, 263–272 (1975)
Felsenstein, J.: Inferring Phylogenies. Sinauer, Sunderland (2004)
Fernandez-Baca, D.: The perfect phylogeny problem. In: Du, D.Z., Cheng, X. (eds.) Steiner Trees in Industries. Kluwer Academic Publishers, Dordrecht (2000)
Fernandez-Baca, D., Lagergren, J.: A polynomial-time algorithm for near-perfect phylogeny. In: Meyer auf der Heide, F., Monien, B. (eds.) ICALP 1996. LNCS, vol. 1099, pp. 670–680. Springer, Heidelberg (1996)
Fitch, W.: Towards finding the tree of maximum parsimony. In: Estabrook, G.F. (ed.) Proceedings of the eighth international conference on numerical taxonomy, pp. 189–230. W.H. Freeman, New York (1975)
Gavril, F.: The intersection graphs of subtrees in trees are exactly the chordal graphs. J. Combinatorial Theory, B 16, 47–56 (1974)
Golumbic, M.C.: Algorithmic Graph Theory and Perfect Graphs. Academic Press, New York (1980)
Gusfield, D.: Efficient algorithms for inferring evolutionary history. Networks 21, 19–28 (1991)
Gusfield, D., Frid, Y., Brown, D.: Integer programming formulations and computations solving phylogenetic and population genetic problems with missing or genotypic data. In: Lin, G. (ed.) COCOON 2007. LNCS, vol. 4598, pp. 51–64. Springer, Heidelberg (2007)
Gusfield, D., Wu, Y.: The three-state perfect phylogeny problem reduces to 2-SAT (to appear)
Heggernes, P.: Minimal triangulations of graphs: A survey. Discrete Mathematics 306, 297–317 (2006)
Hudson, R.: Generating samples under the Wright-Fisher neutral model of genetic variation. Bioinformatics 18(2), 337–338 (2002)
Kannan, S., Warnow, T.: Inferring evolutionary history from DNA sequences. SIAM J. on Computing 23, 713–737 (1994)
Kannan, S., Warnow, T.: A fast algorithm for the computation and enumeration of perfect phylogenies when the number of character states is fixed. SIAM J. on Computing 26, 1749–1763 (1997)
McKee, T.A., McMorris, F.R.: Topics in Intersection Graph Theory. Siam Monographs on Discrete Mathematics (1999)
Parra, A., Scheffler, P.: How to use the minimal separators of a graph for its chordal triangulation. In: Fülöp, Z., Gecseg, F. (eds.) ICALP 1995. LNCS, vol. 944, pp. 123–134. Springer, Heidelberg (1995)
Parra, A., Scheffler, P.: Characterizations and algorithmic applications of chordal graph embeddings. Discrete Applied Mathematics 79, 171–188 (1997)
Pe’er, I., Pupko, T., Shamir, R., Sharan, R.: Incomplete directed perfect phylogeny. SIAM J. on Computing 33, 590–607 (2004)
Semple, C., Steel, M.: Phylogenetics. Oxford University Press, Oxford (2003)
Steel, M.: The complexity of reconstructing trees from qualitative characters and subtrees. J. of Classification 9, 91–116 (1992)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Gusfield, D. (2009). The Multi-State Perfect Phylogeny Problem with Missing and Removable Data: Solutions via Integer-Programming and Chordal Graph Theory. In: Batzoglou, S. (eds) Research in Computational Molecular Biology. RECOMB 2009. Lecture Notes in Computer Science(), vol 5541. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-02008-7_18
Download citation
DOI: https://doi.org/10.1007/978-3-642-02008-7_18
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-02007-0
Online ISBN: 978-3-642-02008-7
eBook Packages: Computer ScienceComputer Science (R0)