Abstract
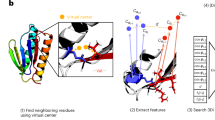
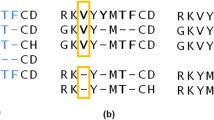
Accurate comparative analysis tools for low-homology proteins remains a difficult challenge in computational biology, especially sequence alignment and consensus folding problems. We presentpartiFold-Align, the first algorithm for simultaneous alignment and consensus folding of unaligned protein sequences; the algorithm’s complexity is polynomial in time and space. Algorithmically,partiFold-Align exploits sparsity in the set of super-secondary structure pairings and alignment candidates to achieve an effectively cubic running time for simultaneous pairwise alignment and folding. We demonstrate the efficacy of these techniques on transmembrane β-barrel proteins, an important yet difficult class of proteins with few known three-dimensional structures. Testing against structurally derived sequence alignments,partiFold-Align significantly outperforms state-of-the-art pairwise sequence alignment tools in the most difficult low sequence homology case and improves secondary structure prediction where current approaches fail. Importantly, partiFold-Align requires no prior training. These general techniques are widely applicable to many more protein families. partiFold-Align is available at http://partiFold.csail.mit.edu.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Shakhnovich, B.E., Deeds, E., Delisi, C., Shakhnovich, E.: Protein structure and evolutionary history determine sequence space topology. Genome Res. 15(3), 385–392 (2005)
Edgar, R.C., Batzoglou, S.: Multiple sequence alignment. Curr. Opin. Struct. Biol. 16(3), 368–373 (2006)
Selbig, J., Mevissen, T., Lengauer, T.: Decision tree-based formation of consensus protein secondary structure prediction. Bioinform. 15(12), 1039–1046 (1999)
Forrest, L.R., Tang, C.L., Honig, B.: On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 91(2), 508–517 (2006)
Sankoff, D.: Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J. Comput. 45(5), 810–825 (1985)
Do, C.B., Foo, C.S., Batzoglou, S.: A max-margin model for efficient simultaneous alignment and folding of RNA sequences. Bioinformatics 24, i68–i76 (2008)
Hofacker, I.L., Bernhart, S.H.F., Stadler, P.F.: Alignment of RNA base pairing probability matrices. Bioinformatics 20(14), 2222–2227 (2004)
Mathews, D.H., Turner, D.H.: Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 317(2), 191–203 (2002)
Havgaard, J.H., Torarinsson, E., Gorodkin, J.: Fast pairwise structural RNA alignments by pruning of the dynamical programming matrix. PLoS Comput. Biol. 3(10), 1896–1908 (2007)
Backofen, R., Will, S.: Local sequence-structure motifs in RNA. J. Bioinform. Comput. Biol. 2(4), 681–698 (2004)
Fariselli, P., Olmea, O., Valencia, A., Casadio, R.: Progress in predicting inter-residue contacts of proteins with neural networks and correlated mutations. Proteins (suppl. 5), 157–162 (2001)
Xu, J., Li, M., Kim, D., Xu, Y.: RAPTOR: Optimal protein threading by linear programming. J. of Bioinform. and Comp. Biol., JBCB (2003)
Bradley, P., Cowen, L., Menke, M., King, J., Berger, B.: Betawrap: Successful prediction of parallel beta-helices from primary sequence reveals an association with many microbial pathogens. Proceedings of the National Academy of Sciences 98(26), 14819–14824 (2001)
Waldispuhl, J., Berger, B., Clote, P., Steyaert, J.M.: Predicting transmembrane beta-barrels and interstrand residue interactions from sequence. Proteins 65(1), 61–74 (2006)
Waldispuhl, J., O’Donnell, C.W., Devadas, S., Clote, P., Berger, B.: Modeling ensembles of transmembrane beta-barrel proteins. Proteins 71(3), 1097–1112 (2008)
Sutormin, R.A., Rakhmaninova, A.B., Gelfand, M.S.: Batmas30: amino acid substitution matrix for alignment of bacterial transporters. Proteins 51, 85–95 (2003)
Henikoff, S., Henikoff, J.: Amino acid substitution matrices from protein blocks. PNAS 89, 10915–10919 (1992)
Rice, P., Longden, I., Bleasby, A.: Emboss: the european molecular biology open software suite. Trends Genet. 16(6), 276–277 (2000)
Will, S., Reiche, K., Hofacker, I.L., Stadler, P.F., Backofen, R.: Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 3(4), e65 (2007)
Caprara, A., Carr, R., Istrail, S., Lancia, G., Walenz, B.: 1001 optimal PDB structure alignments: integer programming methods for finding the maximum contact map overlap. J. Comput. Biol. 11(1), 27–52 (2004)
Lomize, M., Lomize, A., Pogozheva, I., Mosberg, H.: OPM: Orientations of Proteins in Membranes database. Bioinformatics 22, 623–625 (2006)
Menke, M., Berger, B., Cowen, L.: Matt: local flexibility aids protein multiple structure alignment. PLoS Comp. Bio. 4(1), e10 (2008)
Doolittle, R.: Similar amino acid sequences: chance or common ancestry? Science 214, 149–159 (1981)
Raghava, G., Barton, G.: Quantification of the variation in percentage identity for protein sequence alignments. BMC Bioinformatics 7, 415 (2006)
Dunbrack, R.L.J.: Sequence comparison and protein structure prediction. Curr. Opin. Struct. Biol. 16(3), 374–384 (2006)
Cline, M., Hughey, R., Karplus, K.: Predicting reliable regions in protein sequence alignments. Bioinformatics 18(2), 306–314 (2002)
Frishman, D., Argos, P.: Knowledge-based protein secondary structure assignment. Proteins 23, 566–579 (1995)
Edgar, R.C.: Muscle: multiple sequence alignment with high accuracy and high throughput. NAR 32(5), 1792–1797 (2004)
Edgar, R.C.: Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004)
Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., Batzoglou, S.: Lagan and multi-lagan: efficient tools for large-scale multiple alignment of genomic dna. Genome Res. 13(4), 721–731 (2003)
Do, C.B., Woods, D.A., Batzoglou, S.: CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 22(14), e90–e98 (2006)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Waldispühl, J., O’Donnell, C.W., Will, S., Devadas, S., Backofen, R., Berger, B. (2009). Simultaneous Alignment and Folding of Protein Sequences. In: Batzoglou, S. (eds) Research in Computational Molecular Biology. RECOMB 2009. Lecture Notes in Computer Science(), vol 5541. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-02008-7_25
Download citation
DOI: https://doi.org/10.1007/978-3-642-02008-7_25
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-02007-0
Online ISBN: 978-3-642-02008-7
eBook Packages: Computer ScienceComputer Science (R0)