Abstract
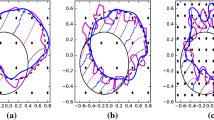
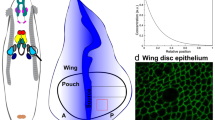
Having acquired images of a growing organism, the question arises how they can be used to infer the properties of growth. We address this challenging image understanding problem by using the GRID (Growth as Random Iterated Diffeomorphisms) model for biological growth. In the GRID model, growth patterns are composed of smaller, local deformations, each resulting from elementary biological events (e.g.,cell division). A large number of such biological events, each occurring randomly and independently from one another, results in a visible growth pattern or biological shape changes as seen in images. A biological transformation underlying observed shape changes is a solution to a GRID visible growth differential equation. We propose its automatic generation via direct estimation of the growth magnitudes from image data. The growth magnitude is a GRID parameter that characterizes a local expansion/contraction rate throughout the organism’s domain. The estimation algorithm is based on the unconstrained optimal control problem formulation expressed in Darcyan coordinates of the organism’s domain and consequent application of Polak-Ribiere minimization routine. We demonstrate the proposed inference method using confocal micrographs of the Drosophila wing disc at larval stage of development.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Bookstein, F.L.: Morphometric Tools for Landmark Data: Geometry and Biology. Cambridge University Press, Cambridge (1991)
Bookstein, F.L.: Biometrics, Biomathematics and the Morphometric Synthesis. Bull. of Math. Biol. 58(2), 313–365 (1996)
Davatzikos, C.: Spatial Transformation and Registration of Brain Images Using Elastically Deformable Models. Comput. Vision Image Under. 66(2), 207–222 (1997)
Kyriacou, S.K., Davatzikos, C., Zinreich, S.J., Bryan, R.N.: Nonlinear Elastic Registration of Brain Images with Tumor Pathology Using Biomechanical Model. IEEE Trans. Med. Imaging 18(7), 580–592 (1999)
Christensen, G.E., Rabbitt, R.D., Miller, M.I.: A Deformable Neuroanatomy Textbook Based on Viscous Fluid Mechanics. In: Proceedings of 1993 Conf. on Information Sciences and Systems, Johns Hopkins University, pp. 211–216 (1999)
Christensen, G.E., Sarang, C.J., Miller, M.I.: Volumetric Transformation of Brain Anatomy. IEEE Trans. Med. Imaging 16(6), 864–876 (1997)
Bookstein, F.L.: Size and Shape Spaces for Landmark Data in Two Dimensions. Statistical Science 1, 181–242 (1998)
Khaneja, N., Miller, M., Grenander, U.: Dynamic Programming Generation of Curves on Brain Surfaces. IEEE Trans. Patt. Anal. Mach. Int. 20(11), 1260–1264 (1998)
Srivastava, A., Saini, S., Ding, Z., Grenander, U.: Maximum-Likelihood Estimation of Biological Growth Variables. In: Rangarajan, A., Vemuri, B.C., Yuille, A.L. (eds.) EMMCVPR 2005. LNCS, vol. 3757, pp. 107–118. Springer, Heidelberg (2005)
Grenander, U.: On the Mathematics of Growth. Quart. Appl. Math. 65, 205–257 (2007)
Grenander, U., Srivastava, A., Saini, S.: A Pattern-Theoretic Characterization of Biological Growth. IEEE Trans. Med. Imaging 26(5), 648–659 (2007)
Portman, N., Grenander, U., Vrscay, E.R.: New computational methods for the construction of “Darcyan” biological coordinate systems. In: Kamel, M.S., Campilho, A. (eds.) ICIAR 2007. LNCS, vol. 4633, pp. 143–156. Springer, Heidelberg (2007)
Carroll, S.B.: Endless Forms Most Beautiful: The New Science of Evo Devo and the Making of the Animal Kingdom. W. W. Norton & Company, NY (2005)
Williams, J.A., Paddock, S.W., Carroll, S.B.: Pattern Formation in a Secondary Field: a Hierarchy of Regulatory Genes Subdivides the Developing Drosophila Wing Disc into Discrete Subregions. Development 117, 571–584 (1993)
Polak, E.: omputational Methods in Optimization: a Unified Approach. Mathematics in Science and Engineering, vol. 77. Academic Press, NY (1971)
Press, W.H., Flannery, B.P., Teukolsky, S.A., Vetterling, W.T.: Numerical Recipes in C: The Art of Scientific Computing. Cambridge University Press, Cambridge (1992)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Portman, N., Grenander, U., Vrscay, E.R. (2009). Direct Estimation of Biological Growth Properties from Image Data Using the “GRID” Model. In: Kamel, M., Campilho, A. (eds) Image Analysis and Recognition. ICIAR 2009. Lecture Notes in Computer Science, vol 5627. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-02611-9_82
Download citation
DOI: https://doi.org/10.1007/978-3-642-02611-9_82
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-02610-2
Online ISBN: 978-3-642-02611-9
eBook Packages: Computer ScienceComputer Science (R0)