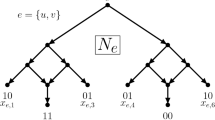
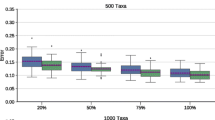
Abstract
Accurate reconstruction of phylogenies remains a key challenge in evolutionary biology. Most biologically plausible formulations of the problem are formally NP-hard, with no known efficient solution. The standard in practice are fast heuristic methods that are empirically known to work very well in general, but can yield results arbitrarily far from optimal. Practical exact methods, which yield exponential worst-case running times but generally much better times in practice, provide an important alternative. We report progress in this direction by introducing a provably optimal method for the weighted multi-state maximum parsimony phylogeny problem. The method is based on generalizing the notion of the Buneman graph, a construction key to efficient exact methods for binary sequences, so as to apply to sequences with arbitrary finite numbers of states with arbitrary state transition weights. We implement an integer linear programming (ILP) method for the multi-state problem using this generalized Buneman graph and demonstrate that the resulting method is able to solve data sets that are intractable by prior exact methods in run times comparable with popular heuristics. Our work provides the first method for provably optimal maximum parsimony phylogeny inference that is practical for multi-state data sets of more than a few characters.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Posada, D., Crandall, K.: Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology and Evolution 16, 37–45 (2001)
Felsenstein, J.: Inferring Phylogenies. Sinauer Publications (2004)
Foulds, L.R., Graham, R.L.: The Steiner problem in phylogeny is NP-complete. Advances in Applied Mathematics 3, 43–49 (1982)
Sridhar, S., Lam, F., Blelloch, G., Ravi, R., Schwartz, R.: Efficiently finding the most parsimonious phylogenetic tree via linear programming. In: Măndoiu, I.I., Zelikovsky, A. (eds.) ISBRA 2007. LNCS (LNBI), vol. 4463, pp. 37–48. Springer, Heidelberg (2007)
Buneman, P.: The recovery of trees from measures of dissimilarity. In: Hodson, F., et al. (eds.) Mathematics in the archeological and historical sciences, pp. 387–395 (1971)
Barthẽlemy, J.: From copair hypergraphs to median graphs with latent vertices. Discrete Math. 76, 9–28 (1989)
Bandelt, H.J., Forster, P., Sykes, B.C., Richards, M.B.: Mitochondrial portraits of human populations using median networks. Genetics 141, 743–753 (1989)
Bandelt, H.J., Forster, P., Rohl, A.: Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16, 37–48 (1999)
Huber, K.T., Moulton, V.: The relation graph. Discrete Mathematics 244(1-3), 153–166 (2002)
Zhou, H.F., Zheng, X.M., Wei, R.X., Second, G., Vaughan, D.A., Ge, S.: Contrasting population genetic structure and gene flow between Oryza rufipogon and Oryza nivara. Theor. Appl. Genet. 117(7), 1181–1189 (2008)
Hudjashov, G., Kivisild, T., Underhill, P.A., Endicott, P., Sanchez, J.J., Lin, A.A., Shen, P., Oefner, P., Renfrew, C., Villems, R., Forster, P.: Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc. Natl. Acad. Sci. USA 104(21), 8726–8730 (2007)
Swofford, D.: PAUP* 4.0. Sinauer Assoc. Inc., Sunderland (2009)
Felsenstein, J.: PHYLIP (phylogeny Inference package) version 3.6 distributed by author, Department of Genome Sciences, University of Washington, Seattle (2008)
Semple, C., Steel, M.: Phylogenetics. Oxford University Press, Oxford (2003)
Erdos, P.L., Szekely, L.A.: On weighted multiway cuts in trees. Mathematical Programming 65, 93–105 (1994)
Wang, L., Jiang, T., Lawler, L.: Approximation algorithms for tree alignment with a given phylogeny. Algorithmica 16, 302–315 (1996)
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Applegate, D.L., Bixby, R.E., Chvatal, V., Cook, W., Espinoza, D.G., Goycoolea, M., Helsgaun, K.: Certification of an optimal TSP tour through 85,900 cities. Operations Research Letters 37(1), 11–15 (2009)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Misra, N., Blelloch, G., Ravi, R., Schwartz, R. (2010). Generalized Buneman Pruning for Inferring the Most Parsimonious Multi-state Phylogeny. In: Berger, B. (eds) Research in Computational Molecular Biology. RECOMB 2010. Lecture Notes in Computer Science(), vol 6044. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-12683-3_24
Download citation
DOI: https://doi.org/10.1007/978-3-642-12683-3_24
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-12682-6
Online ISBN: 978-3-642-12683-3
eBook Packages: Computer ScienceComputer Science (R0)