Abstract
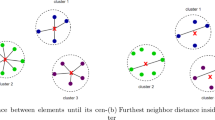
In this article, an evolutionary crisp clustering technique is described that uses a new consensus multiobjective differential evolution. The algorithm is therefore able to optimize two conflicting cluster validity measures simultaneously and provides resultant Pareto optimal set of non-dominated solutions. Thereafter the problem of choosing the best solution from resultant Pareto optimal set is resolved by creation of consensus clusters using voting procedure. The proposed method is used for analyzing the categorical data where no such natural ordering can be found among the elements in categorical domain. Hence no inherent distance measure, like the Euclidean distance, would work to compute the distance between two categorical objects. Index-coded encoding of the cluster medoids (centres) is used for this purpose. The effectiveness of the proposed technique is provided for artificial and real life categorical data sets. Also statistical significance test has been carried out to establish the statistical significance of the clustering results. Matlab version of the software is available at http://bio.icm.edu.pl/~darman/CMODECC.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Jain, A.K., Dubes, R.C.: Algorithms for Clustering Data. Prentice-Hall, Englewood Cliffs (1988)
Kaufman, L., Rousseeuw, P.J.: Finding Groups in Data, An Itroduction to Cluster Analysis. John Wiley and Sons, Brussels (1990)
Storn, R., Price, K.: Differential evolution - A simple and efficient heuristic strategy for global optimization over continuous spaces. Journal of Global Optimization 11, 341–359 (1997)
Maulik, U., Bandyopadhyay, S.: Genetic algorithm based clustering technique. Pattern Recognition 33, 1455–1465 (2000)
Deb, K., Agrawal, S., Pratab, A., Meyarivan, T.: A fast elitist non-dominated sorting genetic algorithm for multi-objective optimization: NSGA-II. IEEE Transactions on Evolutionary Computation 6, 182–197 (2002)
Yeung, K., Ruzzo, W.: An empirical study on principal component analysis for clustering gene expression data. Bioinformatics 17, 763–774 (2001)
Davies, L.D., Bouldin, W.D.: A cluster separation measure. IEEE Transactions on Pattern Analysis and Machine Intelligence 1(2), 224–227 (1979)
Dunn, J.C.: A fuzzy relative of the ISODATA process and its use in detecting compact well-separated clusters. Journal of Cybernetics 3(3), 32–57 (1973)
Vermeulen-Jourdan, L., Dhaenens, C., Talbi, E.G.: Clustering nominal and numerical data: a new distance concept for an hybrid genetic algorithm. In: Gottlieb, J., Raidl, G.R. (eds.) EvoCOP 2004. LNCS, vol. 3004, pp. 220–229. Springer, Heidelberg (2004)
Ferguson, G.A., Takane, Y.: Statistical analysis in psychology and education (2005)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Saha, I., Plewczyński, D., Maulik, U., Bandyopadhyay, S. (2010). Consensus Multiobjective Differential Crisp Clustering for Categorical Data Analysis. In: Szczuka, M., Kryszkiewicz, M., Ramanna, S., Jensen, R., Hu, Q. (eds) Rough Sets and Current Trends in Computing. RSCTC 2010. Lecture Notes in Computer Science(), vol 6086. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-13529-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-642-13529-3_5
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-13528-6
Online ISBN: 978-3-642-13529-3
eBook Packages: Computer ScienceComputer Science (R0)