Abstract
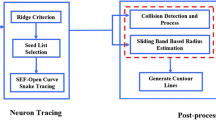
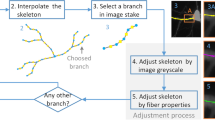
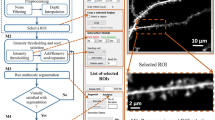
The morphology and structure of 3D dendritic backbones are the essential to understand the neuronal circuitry and behaviors in the neurodegenerative diseases. As a big challenge, the research of extraction of dendritic backbones using image processing and analysis technology has attracted many computational scientists. This paper proposes a reliable and robust approach for automatically extract dendritic backbones in 3D optical microscopy images. Our systematic scheme is a gradient vector field based skeletonization approach. We first use self-snake based nonlinear diffusion, adaptive segmentation to smooth noise and segment the neuron object. Then we propose a hierarchical skeleton points detection algorithm (HSPD) using the measurement criteria of low divergence and high iso-surface principle curvature. We further create a minimum spanning tree to represent and establish effective connections among skeleton points and prune small and spurious branches. To improve the robustness and reliability, the dendrite backbones are refined by B-Spline kernel based data fitting. Experimental results on different datasets demonstrate that our approach has high reliability, good robustness and requires less user interaction.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
London, M., Hausser, M.: Dendritic computation. Annual Review Neuroscience 28(1), 503–532 (2005)
Al-Kofahi, K., Lasek, S., Szarowski, D., Pace, C., Nagy, G., Turner, J.N., Roysam, B.: Rapid Automated Threedimensional Tracing of Neurons from Confocal Image Stacks. IEEE Transactions on Information Technology in Biomedicine 6(2), 171–187
Xu, X., Wong, S.T.C.: Wong, Optical Microscopic Image Processing of Dendritic Spines Morphology. IEEE Signal Processing Magzine 23(4), 132–135 (2006)
Firdaus, J., Kishore, M., Xu, X., Raghu, M., Kun, H., Wong, S.T.C.: Robust 3D reconstruction and identification of dendritic spines from optical microscopy imaging. Medical Image Analysis 13, 167–179 (2009)
Yuan, X., Trachtenberg, J.T., Potter, S.M., Roysam, B.: MDL Constrained 3-D Grayscale Skeletonization Algorithm for Automated Extraction of Dendrites and Spines from Fluorescence Confocal Images. Neuroinformatics 7(4), 213–232 (2009)
Cheng, J., Zhou, X., Miller, E., Witt, R.M., Zhu, J., Sabatini, B.L., Wong, S.T.C.: A novel computational approach for automatic dendrite spines detection in two-photon laser scan microscopy. J. Neurosci Methods 165(1), 122–134 (2007)
Perona, P., Malik, J.: Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Machine Intell. 12(7), 629–639 (1990)
Osher, S., Sethian, J.: Fronts propagating with curvature dependent speed: algorithms based on the Hamilton-Jacobi For Mulation. Journal of Computational Physics 79, 12–49 (1988)
El-Fallah, A.I., Ford, G.E.: The evolution of mean curvature in image filtering. In: Proc. IEEE Internat. Conf. on Image Processing (1), pp. 298–302 (1994)
De Paola, V., Holtmaat, A., Knott, G., Song, S., Wilbrecht, L., Caroni, P., Svoboda, K.: Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 49(6), 780–783 (2006)
Al-Kofahi, Y., Lassoued, W., Lee, W., Roysam, B.: Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Transaction on Biomedical Engineering 57(4), 841–852 (2010)
Boykov, Y., Kolmogorov, V.: Graph cuts and efficient N-D image segmentation. International Journal of Computer Vision 70(2), 109–131 (2006)
Bouix, S., Siddiqi, K.: Divergence-Based Medial Surfaces. In: Vernon, D. (ed.) ECCV 2000. LNCS, vol. 1842, pp. 603–618. Springer, Heidelberg (2000)
Cornea, N.D., Silver, D., Yuan, X., Balasubramanian, R.: Computing hierarchical curve-skeletons of 3D objects. The Visual Computer 21(11), 945–955 (2005)
Davis, M.H., Khotanzad, A., Flamig, D.P., Harms, S.E.: Curvature measurement of 3D objects: evaluation and comparison of three methods. In: International Conference on Image Processing (ICIP 1995), vol. 2, pp. 2627–2631 (1995)
Forssell, L.K., Cohen, S.D.: Using line integral convolution for flow visualization: Curvilinear grids, variable-speed animation, and unsteady flows. IEEE Transactions on Visualizationand Computer Graphics 1(2), 133–141 (1995)
Siek, J.G., Lee, L.-Q., Lumsdaine, A.: The Boost Graph Library: User Guide and Reference Manual, p. 12. Pearson Education Inc., London (2001)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Xiao, L., Yuan, X., Galbreath, Z., Roysam, B. (2010). Automatic and Reliable Extraction of Dendrite Backbone from Optical Microscopy Images. In: Li, K., Jia, L., Sun, X., Fei, M., Irwin, G.W. (eds) Life System Modeling and Intelligent Computing. ICSEE LSMS 2010 2010. Lecture Notes in Computer Science(), vol 6330. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-15615-1_13
Download citation
DOI: https://doi.org/10.1007/978-3-642-15615-1_13
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-15614-4
Online ISBN: 978-3-642-15615-1
eBook Packages: Computer ScienceComputer Science (R0)