Abstract
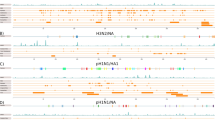
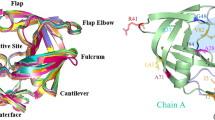
The stability, fold, and the function of proteins need to be maintained throughout the evolution of these molecules – inducing a selective pressure, that can be revealed in sequence data sets. The conservation of structure and function implies coevolution of amino acids within the protein. To understand such selective pressure in the evolution of the human immunodeficiency virus (HIV), we apply information theoretical measures to the two most important enzymes for the progression of viral infection: the reverse transcriptase and the protease. We computed the mutual information to derive insight into the selective pressure acting locally and globally on the enzymes. We found intra- and inter-protein co-evolution of residues in these enzymes and annotate important structural-evolutionary correlations. We discuss a signal indicating a potential co-evolution between the protease and the reverse transcriptase.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Tsygankov, A.Y.: Current developments in anti-HIV/AIDS gene therapy. Curr. Opin. Investig Drugs 10(2), 137–149 (2009)
Wlodawer, A., Erickson, J.: Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62(1), 543–585 (1993)
Perelson, A.S., Neumann, A.U., Markowitz, M., Leonard, J., Ho, D.: HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586 (1996)
Richman, D., Margolis, D., Delaney, M., Greene, W.C., Hazuda, D., Pomerantz, R.J.: The challenge of finding a cure for HIV infection. Science 323, 1304–1307 (2009)
Rong, L., Gilchrist, M.A., Feng, Z., Perelson, A.S.: Modeling within-host HIV-1 dynamics and the evolution of drug resistance: Trade-offs between viral enzyme function and drug susceptibility. J. Theo. Biol. 247, 804–818 (2007)
Chen, L., Lee, C.: Distinguishing HIV-1 drug resistance, accessory, and viral fitness mutations using conditional selection pressure analysis of treated versus untreated patient samples. Biology Direct 1(1), 14 (2006)
Trylska, J., Tozzini, V., Chang, C., McCammon, J.A.: HIV-1 protease substrate binding and product release pathways explored with coarse-grained molecular dynamics. Biophys. J. 92, 4179–4187 (2007)
Hamacher, K., McCammon, J.A.: Computing the amino acid specificity of fluctuations in biomolecular systems. J. Chem. Theory Comput. 2(3), 873–878 (2006)
Hamacher, K.: Relating sequence evolution of HIV1-protease to its underlying molecular mechanics. Gene 422, 30–36 (2008)
Pan, C., Kim, J., Chen, L., Wang, Q., Lee, C.: The hiv positive selection mutation database. Nuc. Acids Res. 35(1), D371–D375 (2007)
Chen, L., Perlina, A., Lee, C.J.: Positive Selection Detection in 40,000 Human Immunodeficiency Virus (HIV) Type 1 Sequences Automatically Identifies Drug Resistance and Positive Fitness Mutations in HIV Protease and Reverse Transcriptase. J. Virol. 78(7), 3722–3732 (2004)
Shannon, C.E.: Prediction and entropy of printed english. The Bell System Technical Journal 30, 50–64 (1951)
Lund, O., Nielsen, M., Lundegaard, C., Brunak, C.K.S.: Immunological Bioinformatics. MIT Press, Cambridge (2005)
Hamacher, K.: Information theoretical measures to analyze trajectories in rational molecular design. J. Comp. Chem. 28(16), 2576–2580 (2007)
Hamacher, K.: Protein domain phylogenies - information theory and evolutionary dynamics. In: Fred, A., Filipe, J., Gamboa, H. (eds.) BIOINFORMATICS 2010, pp. 114–122 (2010)
Pape, S., Hoffgaard, F., Hamacher, K.: Distance-dependent classification of amino acids by information theory. Proteins: Structure, Function, and Bioinformatics 78, 2322–2328 (2010)
Boba, P., Weil, P., Hoffgaard, F., Hamacher, K.: Co-evolution in HIV enzymes. In: Fred, A., Filipe, J., Gamboa, H. (eds.) BIOINFORMATICS 2010, pp. 39–47 (2010)
Weil, P., Hoffgaard, F., Hamacher, K.: Estimating sufficient statistics in co-evolutionary analysis by mutual information. Computational Biology and Chemistry 33(6), 440–444 (2009)
Boba, P., Hamacher, K. (2009), http://bioserver.bio.tu-darmstadt.de/HIV
Press, W.H., et al.: Numerical Recipies in C. Cambridge University Press, Cambridge (1995)
Sarafianos, S.G., Das, K., Hughes, S.H., Arnold, E.: Taking aim at a moving target: designing drugs to inhibit drug-resistant hiv-1 reverse transcriptases. Current Opinion in Structural Biology 14(6), 716–730 (2004)
Prajapati, D.G., Ramajayam, R., Yadav, M.R., Giridhar, R.: The search for potent, small molecule nnrtis: A review. Bioorganic & Medicinal Chemistry 17(16), 5744–5762 (2009)
Yoshimura, K., Kato, R., Yusa, K., Kavlick, M.F., Maroun, V., Nguyen, A., Mimoto, T., Ueno, T., Shintani, M., Falloon, J., Masur, H., Hayashi, H., Erickson, J., Mitsuya, H.: JE-2147: A dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. 96, 8675–8680 (1999)
Reiling, K., Endres, N., Dauber, D., Craik, C., Stroud, R.: Anisotropic dynamics of the JE-2147-HIV protease complex: Drug resistance and thermodynamic binding mode examined in a 1.09 a structure. Biochemistry 41, 4582 (2002)
Perryman, A.L., Lin, J.H., McCammon, J.A.: Restrained molecular dynamics simulations of hiv-1 protease: The first step in validating a new target for drug design. Biopolymers 82(3), 272–284 (2006)
Stone, J.: An Efficient Library for Parallel Ray Tracing and Animation. Master’s thesis, Computer Science Department, University of Missouri-Rolla (April 1998)
Humphrey, W., Dalke, A., Schulten, K.: VMD – Visual Molecular Dynamics. Journal of Molecular Graphics 14, 33–38 (1996)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Boba, P., Weil, P., Hoffgaard, F., Hamacher, K. (2011). Intra- and Inter-Molecular Coevolution: The Case of HIV1 Protease and Reverse Transcriptase. In: Fred, A., Filipe, J., Gamboa, H. (eds) Biomedical Engineering Systems and Technologies. BIOSTEC 2010. Communications in Computer and Information Science, vol 127. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-18472-7_28
Download citation
DOI: https://doi.org/10.1007/978-3-642-18472-7_28
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-18471-0
Online ISBN: 978-3-642-18472-7
eBook Packages: Computer ScienceComputer Science (R0)