Abstract
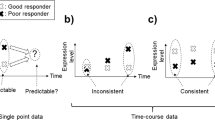
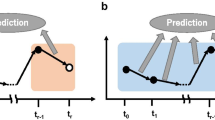
In the last years the constant drive towards a more personalized medicine led to an increasing interest in temporal gene expression analyses. In fact, considering a temporal aspect represents a great advantage to better understand disease progression and treatment results at a molecular level. In this work, we analyse multiple gene expression time series in order to classify the response of Multiple Sclerosis patients to the standard treatment with Interferon-β , to which nearly half of the patients reveal a negative response. In this context, obtaining a highly predictive model of a patient’s response would definitely improve his quality of life, avoiding useless and possibly harmful therapies for the non-responder group. We propose new strategies for time series classification based on biclustering. Preliminary results achieved a prediction accuracy of 94.23% and reveal potentialities to be further explored in classification problems involving other (clinical) time series.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Baranzini, S., Mousavi, P., Rio, J., Stillman, S.C.A., Villoslada, P., Wyatt, M., Comabella, M., Greller, L., Somogyi, R., Montalban, X., Oksenberg, J.: Transcription-based prediction of response to ifnbeta using supervised computational methods. PLoS Biology 3(1) (2005)
Costa, I.G., Schönhuth, A., Hafemeister, C., Schliep, A.: Constrained mixture estimation for analysis and robust classification of clinical time series. Bioinformatics 25(12), i6–i14 (2009)
Hemmer, B., Archelos, J.J., Hartung, H.: New concepts in the immunopathogenesis of multiple sclerosis. Nature Reviews in Neurosciences 3(4), 291–301 (2002)
Lin, T.H., Kaminski, N., Bar-Joseph, Z.: Alignment and classification of time series gene expression in clinical studies. Bioinformatics 24(13), i147–i155 (2008)
Madeira, S.C., Teixeira, M.C., S-Correia, I., Oliveira, A.: Identification of regulatory modules in time series gene expression data using a linear time biclustering algorithm. IEEE/ACM Transactions on Computational Biology and Bioinformatics 7(1), 153–165 (2010)
Sturzebecher, S., Wandinger, K., Rosenwald, A., Sathyamoorthy, M., Tzou, A., Mattar, P., Frank, J., Staudt, L., Martin, R., McFarland, H.: Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain 126(6) (2003)
Ye, L., Keogh, E.: Time series shapelets: a new primitive for data mining. In: Proc. of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (2009)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Carreiro, A.V., Anunciação, O., Carriço, J.A., Madeira, S.C. (2011). Biclustering-Based Classification of Clinical Expression Time Series: A Case Study in Patients with Multiple Sclerosis. In: Rocha, M.P., Rodríguez, J.M.C., Fdez-Riverola, F., Valencia, A. (eds) 5th International Conference on Practical Applications of Computational Biology & Bioinformatics (PACBB 2011). Advances in Intelligent and Soft Computing, vol 93. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-19914-1_31
Download citation
DOI: https://doi.org/10.1007/978-3-642-19914-1_31
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-19913-4
Online ISBN: 978-3-642-19914-1
eBook Packages: EngineeringEngineering (R0)