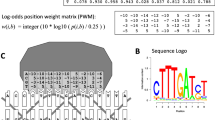
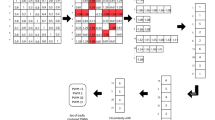
Abstract
Many DNA motif finding algorithms that use Consensus (or any of its variants) in its motif model implicitly impose some restrictive assumptions over transcription factor (TF) binding sites (TFBS). Examples include all binding sites being of equal length, or having exactly one core region with fixed format, etc. In this paper, we have constructed a generalized consensus model (called Mixed-Length-Consensus, or ML-Consensus) without such constraints through multiple sequence alignment of known TFBS. We have extended this model with Information Content (IC) and Pairwise nucleotide correlation Score (PS), and have experimented with using multiple ML-Consensus for a set of binding sites. We have performed leave-one-out cross validation for training and testing of this algorithm over real binding site data of human, mouse, fruit fly, and yeast. We have produced ROC curves (True Positive Rate against False Positive Rate) for these experiments, and have used Wilcoxon Matched-Pair Signed Ranks Test to determine their statistical significance. Our results show that using IC and PS together with ML-Consensus consistently leads to better performance. We have experimented with various scopes for PS, and have found that scope values of 3-5 yield significantly better performance for different configurations. We have also found that using multiple ML-Consensus for one TF significantly improves recognition performance, but single ML-Consensus does better in yeast than in human data. Finally, we have found that using different multiple sequence alignment strategies for ML-Consensus yields varied performance across different species; a naive sorting based multiple sequence alignment outperformed CLUSTAL W2 alignment in yeast data.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Badis, G., et al.: Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 (2009)
Bailey, T.L., Williams, N., Misleh, C., Li, W.W.: MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research 34 (Web Server issue), W369–W373 (2006)
Bulyk, M.L., Johnson, P.L.F., Church, G.M.: Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Research 30(5), 1255–1261 (2002)
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., Frisch, M., Bayerlein, M., Werner, T.: Matinspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21(13), 2933–2942 (2005)
Day, W.H., McMorris, F.: Critical comparison of consensus methods for molecular sequences. Nucleic Acids Research 20(5), 1093–1099 (1992)
Ehret, G., Reichenbach, P., Schindler, U., Horvath, C., Fritz, S., Nabholz, M., Bucher, P.: DNA binding specificity of different STAT proteins. comparison of in vitro specificity with natural target sites. J. Biol. Chem. 276(9), 6675–6688 (2001)
Hannenhalli, S., Wang, L.S.: Enhanced position weight matrices using mixture models. Bioinformatics 21(supplement 1), i204–i212 (2005)
Larkin, M., et al.: Clustal W and Clustal X version 2.0. Bioinformatics 23(21), 2947–2948 (2007)
Lawrence, C.E., Altschul, S.F., Boguski, M.S., Liu, J.S., Neuwald, A.F., Wootton, J.C.: Detecting subtle sequence signals: A gibbs sampling strategy for multiple alignment. Science, New Series 262(5131), 208–214 (1993)
Osada, R., Zaslavsky, E., Singh, M.: Comparative analysis of methods for representing and searching for transcription factor binding sites. Bioinformatics 20(18), 3516–3525 (2004)
Reid, J.E., Evans, K.J., Dyer, N., Wernisch, L., Ott, S.: Variable structure motifs for transcription factor binding sites. BMC Genomics 11(30) (January 14, 2010)
Riley, T., Sontag, E., Chen, P., Levine, A.: Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9(5), 402–412 (2008)
Robison, K., McGuire, A., Church, G.: A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 284, 241–254 (1998)
Sheskin, D.J.: Handbook of Parametric and Nonparametric Statistical Procedures. Chapman & Hall/CRC (2000)
Soldaini, E., John, S., Moro, S., Bollenbacher, J., Schindler, U., Leonard, W.: DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell Biol. 20, 389–401 (2000)
Staden, R.: Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 12, 505–519 (1984)
Stormo, G.D.: DNA binding sites: representation and discovery. Bioinformatics 16(1), 16–23 (2000)
Stormo, G., Fields, D.: Specificity, free energy and information content in protein-DNA interactions. Trends in Biochemical Sciences 23, 109–113 (1998)
Tompa, M., et al.: Assessing computational tools for the discovery of transcription factor binding sites. Nature Biotechnology 23(1), 137–144 (2005)
Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pr, M., Reuter, I., Schacherer, F.: TRANSFAC: an integrated system for gene expression regulation. Nucl. Acids Res. 28(1), 316–319 (2000)
Workman, C.T., Stormo, G.D.: ANN-Spec: a method for discovering transcription factor binding sites with improved specificity. In: Pacific Symposium on Biocomputing, vol. 5, pp. 464–475 (2000)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Quader, S., Snyder, N., Su, K., Mochan, E., Huang, CH. (2011). ML-Consensus: A General Consensus Model for Variable-Length Transcription Factor Binding Sites. In: Pizzuti, C., Ritchie, M.D., Giacobini, M. (eds) Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics. EvoBIO 2011. Lecture Notes in Computer Science, vol 6623. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-20389-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-642-20389-3_3
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-20388-6
Online ISBN: 978-3-642-20389-3
eBook Packages: Computer ScienceComputer Science (R0)