Abstract
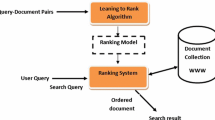
Protein homology prediction is a crucial step in template-based protein structure prediction. The functions that rank the proteins in a database according to their homologies to a query protein is the key to the success of protein structure prediction. In terms of information retrieval, such functions are called ranking functions, and are often constructed by machine learning approaches. Different from traditional machine learning problems, the feature vectors in the ranking-function learning problem are not identically and independently distributed, since they are calculated with regard to queries and may vary greatly in statistical characteristics from query to query. At present, few existing algorithms make use of the query-dependence to improve ranking performance. This paper proposes a query-adaptive ranking-function learning algorithm for protein homology prediction. Experiments with the support vector machine (SVM) used as the benchmark learner demonstrate that the proposed algorithm can significantly improve the ranking performance of SVMs in the protein homology prediction task.
This work was supported by the Research Initiation Funds for President Scholarship Winners of Chinese Academy of Sciences (CAS), the National Natural Science Foundation of China (30900262, 61003140 and 61033010), the CAS Knowledge Innovation Program (KGGX1-YW-13), and the Fundamental Research Funds for the Central Universities (09lgpy62).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Baeza-Yates, R., Ribeiro-Neto, B.: Modern Information Retrieval. Addison-Wesley-Longman, Harlow (1999)
Robertson, S.E., Sparck Jones, K.: Relevance weighting of search terms. Journal of American Society for Information Sciences 27, 129–146 (1976)
Fuhr, N.: Optimal polynomial retrieval functions based on the probability ranking principle. ACM Transactions on Information Systems 7, 183–204 (1989)
Cohen, W., Shapire, R., Singer, Y.: Learning to order things. Journal of Artificial Intelligence Research 10, 243–270 (1999)
Joachims, T.: Optimizing Search Engines Using Clickthrough Data. In: 8th ACM Conference on Knowledge Discovery and Data Mining, pp. 133–142. ACM Press, New York (2002)
Baker, D., Sali, A.: Protein structure prediction and structural genomics. Science 294, 93–96 (2001)
Zhang, Y., Skolnick, J.: The protein structure prediction problem could be solved using the current PDB library. Proc. Natl. Acad. Sci. USA 102, 1029–1034 (2005)
Ginalski, K.: Comparative modeling for protein structure prediction. Current Opinion in Structural Biology 16, 172–177 (2006)
Zhang, Y.: Progress and challenges in protein structure prediction. Current Opinion in Structural Biology 18, 342–348 (2008)
Soding, J.: Protein homology detection by HMMCHMM comparison. Bioinformatics 2, 951–960 (2005)
Teodorescu, O., Galor, T., Pillardy, J., Elber, R.: Enriching the sequence substitution matrix by structural information. Proteins: Structure, Function and Bioinformatics 54, 41–48 (2004)
Cooper, W., Gey, F., Chen, A.: Information retrieval from the TIPSTER collection: an application of staged logistic regression. In: 1st NIST Text Retrieval Conference, pp. 73–88. National Institute for Standards and Technology, Washington, DC (1993)
Gey, F.: Inferring Probability of Relevance Using the Method of Logistic Regression. In: 17th Annual International ACM Conference on Research and Development in Information Retrieval, Dublin, Ireland, pp. 222–231 (1994)
Nallapati, R.: Discriminative Models for Information Retrieval. In: 27th Annual International ACM Conference on Research and Development in Information Retrieval, pp. 64–71. ACM Press, New York (2004)
Herbrich, R., Obermayer, K., Graepel, T.: Large margin rank boundaries for ordinal regression. In: Smola, A.J., Bartlett, P., Schölkopf, B., Schuurmans, C. (eds.) Advances in Large Margin Classifiers, pp. 115–132. MIT Press, Cambridge (2000)
Crammer, K., Singer, Y.: Pranking with ranking. In: Advances in Neural Information Processing Systems, vol. 14, pp. 641–647. MIT Press, Cambridge (2002)
Chapelle, O., Keerthi, S.S.: Efficient algorithms for ranking with SVMs. Information Retrieval Journal 13, 201–215 (2010)
McFee, B., Lanckriet, G.: Metric Learning to Rank. In: 27th International Conference on Machine Learning, Haifa, Israel (2010)
Fu, Y., Sun, R., Yang, Q., He, S., Wang, C., Wang, H., Shan, S., Liu, J., Gao, W.: A Block-Based Support Vector Machine Approach to the Protein Homology Prediction Task in KDD Cup 2004. SIGKDD Explorations 6, 120–124 (2004)
Fu, Y.: Machine Learning Based Bioinformation Retrieval. Ph.D. Thesis, Institute of Computing Technology, Chinese Academy of Sciences (2007)
Vapnik, V.N.: The Nature of Statistical Learning Theory. Springer, New York (1995)
Foussette, C., Hakenjos, D., Scholz, M.: KDD-Cup 2004 - Protein Homology Task. SIGKDD Explorations 6, 128–131 (2004)
Pfahringer, B.: The Weka Solution to the 2004 KDD Cup. SIGKDD Explorations 6, 117–119 (2004)
Tang, Y., Jin, B., Zhang, Y.: Granular Support Vector Machines with Association Rules Mining for Protein Homology Prediction. Special Issue on Computational Intelligence Techniques in Bioinformatics, Artificial Intelligence in Medicine 35, 121–134 (2005)
Caruana, R., Joachims, T., Backstrom, L.: KDD Cup 2004: Results and Analysis. SIGKDD Explorations 6, 95–108 (2004)
Tobi, D., Elber, R.: Distance dependent, pair potential for protein folding: Results from linear optimization. Proteins, Structure Function and Genetics 41, 16–40 (2000)
Joachims, T.: Making large-Scale SVM Learning Practical. In: Schölkopf, B., Burges, C., Smola, A. (eds.) Advances in Kernel Methods - Support Vector Learning, pp. 115–132. MIT Press, Cambridge (1999)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Fu, Y., Pan, R., Yang, Q., Gao, W. (2011). Query-Adaptive Ranking with Support Vector Machines for Protein Homology Prediction. In: Chen, J., Wang, J., Zelikovsky, A. (eds) Bioinformatics Research and Applications. ISBRA 2011. Lecture Notes in Computer Science(), vol 6674. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-21260-4_31
Download citation
DOI: https://doi.org/10.1007/978-3-642-21260-4_31
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-21259-8
Online ISBN: 978-3-642-21260-4
eBook Packages: Computer ScienceComputer Science (R0)