Abstract
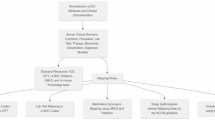
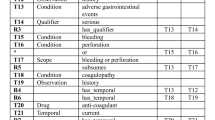
The completion of clinical trial depends on sufficient participant enrollment, which is often problematic due to the restrictiveness of eligibility criteria, and effort required to verify patient eligibility. The objective of this research is to support the design of eligibility criteria, enable the reuse of structured criteria and to provide meaningful suggestions of relaxing them based on previous trials. The paper presents the first steps, a method for automatic comparison of criteria content and the library of structured and ordered eligibility criteria that can be browsed with the fine-grained queries. The structured representation consists of the automatically identified contextual patterns and semantic entities. The comparison of criteria is based on predefined relations between the patterns, concept equivalences defined in medical ontologies, and finally on threshold values. The results are discussed from the perspective of the scope of the eligibility criteria covered by our library.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Cuggia, M., Besana, P., Glasspool, D.: Comparing semi-automatic systems for recruitment of patients to clinical trials. International Journal of Medical Informatics 80(6), 371–388 (2011)
Milian, K., ten Teije, A., Bucur, A., van Harmelen, F.: Patterns of Clinical Trial Eligibility Criteria. In: Riaño, D., ten Teije, A., Miksch, S. (eds.) KR4HC 2011. LNCS, vol. 6924, pp. 145–157. Springer, Heidelberg (2012)
Cunningham, H., Maynard, D., Bontcheva, K., Tablan, V.: GATE: A framework and graphical development environment for robust NLP tools and applications. In: Proceedings of the 40th Anniversary Meeting of the ACL (2002)
Aronson, A.R.: Metamap: Mapping text to the umls metathesaurus. In: Proceedings AMIA Symposium (2001)
G. Units, Units (November 2006), http://www.gnu.org/software/units/
Hassanzadeh, O., Kementsietsidis, A., Lim, L., Miller, R.J., Wang, M.: Linkedct: A linked data space for clinical trials, CoRR abs/0908.0567
Wang, S., Ohno-Machado, L., Mar, P., Boxwala, A., Greenes, R.: Enhancing arden syntax for clinical trial eligibility criteria. In: Proceedings AMIA Symposium
Sordo, M., Ogunyemi, O., Boxwala, A.A., Greenes, R.A.: Gello: An object-oriented query and expression language for clinical decision support. In: Proceedings AMIA Symposium, vol. (5), p. 1012 (2003)
Tu, S., Peleg, M., Carini, S., Rubin, D., Sim, I.: Ergo: A templatebased expression language for encoding eligibility criteria. Tech. rep. (2009)
Weng, C., Tu, S.W., Sim, I., Richesson, R.: Formal representations of eligibility criteria: A literature review. Journal of Biomedical Informatics (2009)
Tu, S., Peleg, M., Carini, S., Bobak, M., Rubin, D., Sim, I.: A practical method for transforming free-text eligibility criteria into computable criteria
Cheng, X.-y., Chen, X.-h., Hua, J.: The overview of entity relation extraction methods. Intelligent Computing and Information Science 134, 749–754 (2011)
Nammuni, K., Pickering, C., Modgil, S., Montgomery, A., Hammond, P., Wyatt, J.C., Altman, D.G., Dunlop, R., Potts, H.W.W.: Design-a-trial: a rule-based decision support system for clinical trial design. Knowledge-Based Systems
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Milian, K., Bucur, A., van Harmelen, F. (2012). Building a Library of Eligibility Criteria to Support Design of Clinical Trials. In: ten Teije, A., et al. Knowledge Engineering and Knowledge Management. EKAW 2012. Lecture Notes in Computer Science(), vol 7603. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-33876-2_29
Download citation
DOI: https://doi.org/10.1007/978-3-642-33876-2_29
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-33875-5
Online ISBN: 978-3-642-33876-2
eBook Packages: Computer ScienceComputer Science (R0)