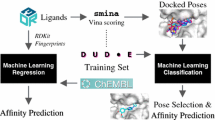
Abstract
Accurately predicting the binding affinities of large sets of protein-ligand complexes efficiently is a key challenge in computational biomolecular science, with applications in drug discovery, chemical biology, and structural biology. Since a scoring function (SF) is used to score, rank, and identify potential drug leads, the fidelity with which it predicts the affinity of a ligand candidate for a protein’s binding site has a significant bearing on the accuracy of virtual screening. Despite intense efforts in developing conventional SFs, which are either force-field based, knowledge-based, or empirical, their limited scoring and ranking accuracies have been a major roadblock toward cost-effective drug discovery. Therefore, in this work, we examine a range of SFs employing different machine-learning (ML) approaches in conjunction with a variety of physicochemical and geometrical features characterizing protein-ligand complexes. We compare the scoring and ranking accuracies of these ML SFs as well as those of conventional SFs in the context of the diverse test sets of the 2007 and 2010 PDBbind benchmarks. We also investigate the influence of the size of the training dataset and the number of features used on scoring and ranking accuracies. We find that the best performing ML SF has a scoring power of 0.807 in terms of Pearson correlation coefficient between predicted and measured binding affinities compared to 0.644 achieved by a state-of-the-art conventional SF. Despite this substantial improvement (25%) in binding affinity prediction, the ranking power improvement is only 6% from a success rate of 58.5% achieved by the best conventional SF to 62.2% obtained by the best ML approach when ligands were ranked for 65 unique proteins.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Ewing, T.J.A., Makino, S., Skillman, A.G., Kuntz, I.D.: Dock 4.0: search strategies for automated molecular docking of flexible molecule databases. Journal of Computer-Aided Molecular Design 15(5), 411–428 (2001)
Wang, R., Lai, L., Wang, S.: Further development and validation of empirical scoring functions for structure-based binding affinity prediction. Journal of Computer-Aided Molecular Design 16, 11–26 (2002), doi:10.1023/A:1016357811882
Gohlke, H., Hendlich, M., Klebe, G.: Knowledge-based scoring function to predict protein-ligand interactions. Journal of Molecular Biology 295(2), 337 (2000)
Cheng, T., Li, X., Li, Y., Liu, Z., Wang, R.: Comparative assessment of scoring functions on a diverse test set. Journal of Chemical Information and Modeling 49(4), 1079–1093 (2009)
Ashtawy, H.M., Mahapatra, N.R.: A comparative assessment of conventional and machine-learning-based scoring functions in predicting binding affinities of protein-ligand complexes. In: 2011 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 627–630. IEEE (2011)
Ashtawy, H.M., Mahapatra, N.R.: A comparative assessment of ranking accuracies of conventional and machine-learning-based scoring functions for protein-ligand binding affinity prediction. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 9(5), 1301–1313 (2012)
Ballester, P., Mitchell, J.: A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 26(9), 1169 (2010)
Wang, R., Fang, X., Lu, Y., Wang, S.: The PDBbind database: Collection of binding affinities for protein-ligand complexes with known three-dimensional structures. Journal of Medicinal Chemistry 47(12), 2977–2980 (2004); PMID: 15163179
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E.: The protein data bank. Nucleic Acids Research 28(1), 235–242 (2000)
Madden, T.: The blast sequence analysis tool. The NCBI Handbook. National Library of Medicine (US), National Center for Biotechnology Information (2002)
Schnecke, V., Kuhn, L.A.: Virtual screening with solvation and ligand-induced complementarity. In: Klebe, G. (ed.) Virtual Screening: An Alternative or Complement to High Throughput Screening? pp. 171–190. Springer, Netherlands (2002)
Accelrys Inc., The Discovery Studio Software, San Diego, CA (2001) (version 2.0)
Tripos, Inc., The SYBYL Software, 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA (2006) (version 7.2)
Jones, G., Willett, P., Glen, R., Leach, A., Taylor, R.: Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology 267(3), 727–748 (1997)
Schrödinger, L.: The Schrödinger Software, New York (2005) (version 8.0)
Velec, H.F.G., Gohlke, H., Klebe, G.: DrugScore CSD - knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. Journal of Medicinal Chemistry 48(20), 6296–6303 (2005)
Hastie, T., Tibshirani, R., Friedman, J.: The elements of statistical learning (2001)
R Development Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2010)
Stephen Milborrow, T.H., Tibshirani, R.: earth: Multivariate Adaptive Regression Spline Models (2010) (R package version 2.4-5)
Hechenbichler, K.S.K.: Kknn: Weighted k-Nearest Neighbors (2010) (R package version 1.0-8)
Dimitriadou, E., Hornik, K., Leisch, F., Meyer, D., Weingessel, A.: e1071: Miscellaneous Functions of the Department of Statistics (e1071), TU Wien (2010) (R package version 1.5-24)
Breiman, L.: Random forests. Machine Learning 45, 5–32 (2001)
Ridgeway, G.: Gbm: Generalized Boosted Regression Models (2010) (R package version 1.6-3.1)
Breiman, L.: Bias, variance, and arcing classifiers (technical report 460). Statistics Department, University of California (1996)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Ashtawy, H.M., Mahapatra, N.R. (2013). Does Accurate Scoring of Ligands against Protein Targets Mean Accurate Ranking?. In: Cai, Z., Eulenstein, O., Janies, D., Schwartz, D. (eds) Bioinformatics Research and Applications. ISBRA 2013. Lecture Notes in Computer Science(), vol 7875. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-38036-5_29
Download citation
DOI: https://doi.org/10.1007/978-3-642-38036-5_29
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-38035-8
Online ISBN: 978-3-642-38036-5
eBook Packages: Computer ScienceComputer Science (R0)