Abstract
At sites of chronic inflammation caused by persistent microbial infection or foreign objects, macrophages can fuse forming multinucleated giant cells. On implanted materials, these foreign body-type giant cells (FBGC) are associated with material degradation and device failure. Thus, formation of multinucleated cells is of relevance for the biomaterials field, particularly for the design of novel strategies for bone regeneration. After cell differentiation on varying conditions, multinucleation is analyzed by an independent observer and the percentage of fusion quantified. However, this analysis is still mostly performed manually, being time consuming and prone to errors.
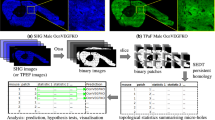
We propose a fully automated method for nuclei detection and the nuclei groups formed in each giant cell. To perform such analysis with robustness and increased performance we use the Laplacian of Gaussian local image filter for cell nuclei detection and Delaunay graphs for the analysis of the formation of nuclei groups characteristic of giant multinucleated cells. Nuclei groups are detected based on both distance between cell nuclei and the properties of the cytoplasm between them.
Results show a good performance in nuclei detection in these images (87.6%), with reasonable error level in giant cell detection (22%), indicating some difficulty in analyzing this data.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Anderson, J.M.: Multinucleated giant cells. Curr. Opin. Hematol. 7, 40–47 (2000)
McNally, A.K., Anderson, J.M.: Foreign body-type multinucleated giant cells induced by interleukin-4 express select lymphocyte co-stimulatory molecules and are phenotypically distinct from osteoclasts and dendritic cells. Experimental and Molecular Pathology 91, 673–681 (2011)
Wiggins, M.J., Wilkoff, B., Anderson, J.M., Hiltner, A.: Biodegradation of polyether polyurethane inner insulation in bipolar pacemaker leads. J. Biomed. Mater. Res. 58, 302–307 (2001)
Anderson, J.M.: Inflammatory response to implants. Am. Soc. Artif. Intern. Organs 11, 101–107 (1988)
Meijering, E.: Cell segmentation: 50 years down the road (life sciences). IEEE Signal Processing Magazine 29(5), 140–145 (2012)
Pang, B., Zhou, L., Zeng, W., You, X.: Cell nuclei detection in histopathological images by using multi-curvature edge cue. In: 2010 IEEE International Conference on Computational Intelligence and Security, pp. 1095–1099 (2011)
Marcuzzo, M., Quelhas, P., Campilho, A., Mendonça, A.M., Campilho, A.: Automated arabidopsis plant root cell segmentation based on svm classification and region merging. Computers in Biology and Medicine 39(9) (2009)
Usaj, M., Torkar, D., Kanduser, M., Mikalavcic, D.: Cell counting tool parameters optimization approach for electroporation efficiency determination of attached cells in phase contrast images. J. of Microscopy 241(3), 303–314 (2011)
Esteves, T., Quelhas, P., Mendonça, A.M., Campilho, A.: Gradient convergence filters and a phase congruency approach for in vivo cell nuclei detection. Machine Vision and Applications 23, 623–638 (2012)
Chen, Y., Quelhas, P., Campilho, A.: Low frame rate cell tracking: A delaunay graph matching approach. In: Proc. of the IEEE Int. conference on Biomedical imaging (ISBI), pp. 1–4 (2011)
Leal, P., Ferro, L., Marques, M., Romão, S., Cruz, T., Tomá, A.M., Castro, H., Quelhas, P.: Automatic assessment of leishmania infection indexes on in vitro macrophage cell cultures. In: Campilho, A., Kamel, M. (eds.) ICIAR 2012, Part II. LNCS, vol. 7325, pp. 432–439. Springer, Heidelberg (2012)
Quelhas, P., Mendonça, A.M., Campilho, A.: 3d cell nuclei fluorescence quantification using sliding band filter. In: International Conference on Pattern Recognition (ICPR), pp. 2508–2511 (2010)
Wen, Q., Chang, H., Parvin, B.: A delaunay triangulation approach for segmenting clumps of nuclei. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp. 9–12 (July 2009)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Ferro, L. et al. (2013). Multinuclear Cell Analysis Using Laplacian of Gaussian and Delaunay Graphs. In: Sanches, J.M., Micó, L., Cardoso, J.S. (eds) Pattern Recognition and Image Analysis. IbPRIA 2013. Lecture Notes in Computer Science, vol 7887. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-38628-2_52
Download citation
DOI: https://doi.org/10.1007/978-3-642-38628-2_52
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-38627-5
Online ISBN: 978-3-642-38628-2
eBook Packages: Computer ScienceComputer Science (R0)