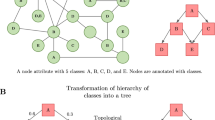
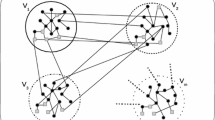
Abstract
We present an algorithm for hierarchical multi-label classification (HMC) in a network context. It is able to classify instances that may belong to multiple classes at the same time and consider the hierarchical organization of the classes. It assumes that the instances are placed in a network and uses information on the network connections during the learning of the predictive model. Many real world prediction problems have classes that are organized hierarchically and instances that can have pairwise connections. One example is web document classification, where topics (classes) are typically organized into a hierarchy and documents are connected by hyperlinks. Another example, which is considered in this paper, is gene/protein function prediction, where genes/proteins are connected and form protein-to-protein interaction (PPI) networks. Network datasets are characterized by a form of autocorrelation, where the value of a variable at a given node depends on the values of variables at the nodes it is connected with. Combining the hierarchical multi-label classification task with network prediction is thus not trivial and requires the introduction of the new concept of network autocorrelation for HMC. The proposed algorithm is able to profitably exploit network autocorrelation when learning a tree-based prediction model for HMC. The learned model is in the form of a Predictive Clustering Tree (PCT) and predicts multiple (hierarchically organized) labels at the leaves. Experiments show the effectiveness of the proposed approach for different problems of gene function prediction, considering different PPI networks. The results show that different networks introduce different benefits in different problems of gene function prediction.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Ashburner, M., et al.: Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genetics 25, 25–29 (2000)
Astikainen, K., Pitkänen, E., Rousu, J., Holm, L., Szedmák, S.: Reaction kernels - structured output prediction approaches for novel enzyme function. Bioinformatics, 48–55 (2010)
Barutcuoglu, Z., Schapire, R.E., Troyanskaya, O.G.: Hierarchical multi-label prediction of gene function. Bioinformatics 22(7), 830–836 (2006)
Batagelj, V., Mrvar, A.: PAJEK – Program for large network analysis (1998)
Bi, W., Kwok, J.T.: Multilabel classification on tree- and dag-structured hierarchies. In: Getoor, L., Scheffer, T. (eds.) ICML, pp. 17–24. Omnipress (2011)
Bilgic, M., Getoor, L.: Effective label acquisition for collective classification. In: Proc. 14th ACM SIGKDD Intl. Conf on Knowledge Discovery and Data Mining, pp. 43–51 (2008)
Ceci, M.: Hierarchical text categorization in a transductive setting. In: ICDM Workshops, pp. 184–191 (2008)
Ceci, M., Malerba, D.: Classifying web documents in a hierarchy of categories: a comprehensive study. J. Intell. Inf. Syst. 28(1), 37–78 (2007)
Cerri, R., Barros, R.C., de Carvalho, A.C.P.L.F.: A genetic algorithm for hierarchical multi-label classification. In: Proc. of the 27th Annual ACM Symposium on Applied Computing, SAC 2012, pp. 250–255. ACM (2012)
Cesa-Bianchi, N., Gentile, C., Zaniboni, L.: Incremental algorithms for hierarchical classification. J. Mach. Learn. Res. 7, 31–54 (2006)
Clare, A., King, R.D.: Predicting gene function in s. cerevisiae. In: Proc. Eur. Conf. on Computational Biology, pp. 42–49 (2003)
Deane, C.M., Salwiński, Ł., Xenarios, I., Eisenberg, D.: Protein interactions. Molecular & Cellular Proteomics: MCP 1(5), 349–356 (2002)
Doreian, P.: Network Autocorrelation Models: Problems and Prospects. In: Spatial Statistics: Past, Present, and Future. Monograph, vol. 12. Ann Arbor Institute of Mathematical Geography (1990)
Gallagher, B., Tong, H., Eliassi-Rad, T., Faloutsos, C.: Using ghost edges for classification in sparsely labeled networks. In: Proc. 14th ACM SIGKDD Intl. Conf. Knowledge Discovery and Data Mining, pp. 256–264 (2008)
Jensen, D., Neville, J.: Linkage and autocorrelation cause feature selection bias in relational learning. In: Proc. 9th Intl. Conf. on Machine Learning, pp. 259–266. Morgan Kaufmann (2002)
Jensen, D., Neville, J., Gallagher, B.: Why collective inference improves relational classification. In: Proc. 10th Intl. Conf. on Knowledge Discovery and Data Mining, pp. 593–598 (2004)
Jiang, X., Nariai, N., Steffen, M., Kasif, S., Kolaczyk, E.: Integration of relational and hierarchical network information for protein function prediction. BMC Bioinformatics 9(1) (2008)
Kong, X., Shi, X., Yu, P.S.: Multi-label collective classification. In: SDM, pp. 618–629. SIAM/Omnipress (2011)
Macskassy, S., Provost, F.: Classification in networked data: a toolkit and a univariate case study. Machine Learning 8, 935–983 (2007)
Macskassy, S.A.: Improving learning in networked data by combining explicit and mined links. In: Proc. 22nd Intl. Conf. on Artificial Intelligence, pp. 590–595 (2007)
Mewes, H.W., Heumann, K., Kaps, A., Mayer, K., Pfeiffer, F., Stocker, S., Frishman, D.: Mips: A database for protein sequences and complete genomes. Nucl. Acids Res. 27, 44–48 (1999)
Neville, J., Jensen, D.: Relational dependency networks. Journal of Machine Learning Research 8, 653–692 (2007)
Rahmani, H., Blockeel, H., Bender, A.: Predicting the functions of proteins in protein-protein interaction networks from global information. Journal of Machine Learning Research 8, 82–97 (2010)
Re, M., Valentini, G.: An experimental comparison of hierarchical bayes and true path rule ensembles for protein function prediction. In: El Gayar, N., Kittler, J., Roli, F. (eds.) MCS 2010. LNCS, vol. 5997, pp. 294–303. Springer, Heidelberg (2010)
Rousu, J., Saunders, C., Szedmak, S., Shawe-Taylor, J.: Kernel-based learning of hierarchical multilabel classification models. J. Mach. Learn. Res. 7, 1601–1626 (2006)
Ruepp, et al.: The funcat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Research 32(18), 5539–5545 (2004)
Sen, P., Namata, G., Bilgic, M., Getoor, L., Gallagher, B., Eliassi-Rad, T.: Collective classification in network data. AI Magazine 3, 93–106 (2008)
Steinhaeuser, K., Chawla, N.V., Ganguly, A.R.: Complex networks as a unified framework for descriptive analysis and predictive modeling in climate science. Statistical Analysis and Data Mining 4(5), 497–511 (2011)
Stojanova, D., Ceci, M., Appice, A., Džeroski, S.: Network regression with predictive clustering trees. Data Mining and Knowledge Discovery 14 (2012)
Valentini, G.: True path rule hierarchical ensembles for genome-wide gene function prediction. IEEE ACM Transactions on Computational Biology and Bioinformatics 8(3), 832–847 (2010)
Vens, C., Struyf, J., Schietgat, L., Džeroski, S., Blockeel, H.: Decision trees for hierarchical multi-label classification. Machine Learning 73(2), 185–214 (2008)
von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., Bork, P.: Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417(6887), 399–403 (2002)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Stojanova, D., Ceci, M., Malerba, D., Džeroski, S. (2013). Learning Hierarchical Multi-label Classification Trees from Network Data. In: Fürnkranz, J., Hüllermeier, E., Higuchi, T. (eds) Discovery Science. DS 2013. Lecture Notes in Computer Science(), vol 8140. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-40897-7_16
Download citation
DOI: https://doi.org/10.1007/978-3-642-40897-7_16
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-40896-0
Online ISBN: 978-3-642-40897-7
eBook Packages: Computer ScienceComputer Science (R0)