Abstract
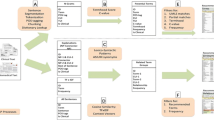
In this paper, we describe the use of ontologies in the context of a system for identifying patients that are eligible for clinical trials. The main purpose of this clinical research data warehouse (CRDW) is to support patient recruitment based on routine data from the hospital’s clinical information system (CIS). In contrast to most other systems for similar purposes, the CRDW also makes use of information present in clinical documents like admission reports, radiological findings and discharge letters. The linguistic analysis recognizes negated and coordinated phrases. It is supported by clinical domain ontologies that enable the identification of main terms and their properties, as well as semantic search with synonyms, hypernyms, and syntactic variants. The CRDW system is currently being tested at hospitals of the Charité - Universitätsmedizin Berlin and the Vivantes - Netzwerk für Gesundheit GmbH. In the paper, we provide an evaluation of the system based on real world data obtained from the daily routine work of the study assistants.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Bodenreider, O.: The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Research 32(Database-Issue), 267–270 (2004)
Browne, P.: Jboss Drools Business Rules. From technologies to solutions. Packt Publishing, Limited (2009)
Chapman, W.W., Hilert, D., Velupillai, S., Kvist, M., Skeppsted, M., Chapman, B.E., Conway, M., Tharp, M., Mowery, D.L., Deleger, L.: Extending the negex lexicon for multiple languages. In: Proceedings of the 14th World Congress on Medical and Health Informatics, MEDINFO 2013 (2013)
Cowie, J., Wilks, Y.: Information extraction. In: Handbook of Natural Language Processing, pp. 241–260 (2000)
Cunningham, H., Tablan, V., Roberts, A., Bontcheva, K.: Getting more out of biomedical documents with gate’s full lifecycle open source text analytics
Dugas, M., Lange, M., Berdel, W., Müller-Tidow, C.: Workflow to improve patient recruitment for clinical trials within hospital information systems - a case-study. Trials 9(1), 2 (2008)
Gallaire, H., Minker, J., Nicolas, J.-M.: Logic and databases: A deductive approach. ACM Comput. Surv. 16(2), 153–185 (1984)
Jurafsky, D., Martin, J.H.: Speech and Language Processing, 2nd edn. Prentice Hall Series in Artificial Intelligence. Prentice Hall (May 2008)
Kifer, M.: Rule interchange format: The framework. In: Calvanese, D., Lausen, G. (eds.) RR 2008. LNCS, vol. 5341, pp. 1–11. Springer, Heidelberg (2008)
Kifer, M., Lausen, G., Wu, J.: Logical foundations of object-oriented and frame-based languages. Journal of the ACM 42(4), 741–843 (1995)
Lloyd, J.W.: Foundations of Logic Programming, 2nd edn. Springer (1987)
Müller, F.: A finite-state approach to shallow parsing and grammatical functions annotation of German. PhD thesis, University of Tübingen (2005)
Murphy, S.N., Mendis, M.E., Berkowitz, D.A., Kohane, I., Chueh, H.: Integration of clinical and genetic data in the i2b2 architecture. In: AMIA Annu. Symp. Proc., p. 2009 (2006)
Polleres, A.: From SPARQL to rules (and back). In: Williamson, C.L., Zurko, M.E., Patel-Schneider, P.F., Shenoy, P.J. (eds.) WWW, pp. 787–796. ACM (2007)
Reeve, L.: Survey of semantic annotation platforms. In: Proceedings of the 2005 ACM Symposium on Applied Computing, pp. 1634–1638. ACM Press (2005)
Rogers, F.B.: Medical subject headings. Bull. Med. Libr. Assoc. 51, 114–116 (1963)
Rosse, C., Mejino, J.V.L.: A reference ontology for biomedical informatics: the foundational model of anatomy. J. Biomed. Inform. 36, 478–500 (2003)
Ruch, P., Gobeill, J., Lovis, C., Geissbühler, A.: Automatic medical encoding with SNOMED categories. BMC Medical Informatics and Making 8, 6 (2008)
Russell, S.J., Norvig, P.: Artificial Intelligence: A Modern Approach, 2nd edn. Pearson Education (2003)
Scheitz, J.F., Mochmann, H.C., Witzenbichler, B., Fiebach, B., Audebert, H.J., Nolte, C.H.: J. Neurol. 25 (2012)
Scheitz, J.F., Mochmann, H.C., Nolte, C.H., Haeusler, K.G., Audebert, H.J., Heuschmann, P.U., Laufs, U., Witzenbichler, B., Schultheiss, H.P., Endres, M.: Troponin elevation in acute ischemic stroke (TRELAS) – protocol of a prospective observational trial. M. BMC Neurol. 11(98) (2011)
Schwaber, K., Beedle, M.: Agile Software Development with Scrum, 1st edn. Prentice Hall PTR, Upper Saddle River (2001)
Shvaiko, P., Euzenat, J.: Ontology matching: State of the art and future challenges. IEEE TKDE 99 (2011)
Staab, S., Studer, R.: Handbook on Ontologies, 2nd edn. Springer (2009)
Szarvas, G., Farkas, R., Busa-Fekete, R.: Research paper: State-of-the-art anonymization of medical records using an iterative machine learning framework. JAMIA 14(5), 574–580 (2007)
Todorov, K., Geibel, P., Kühnberger, K.-U.: Mining concept similarities for heterogeneous ontologies. In: Perner, P. (ed.) ICDM 2010. LNCS, vol. 6171, pp. 86–100. Springer, Heidelberg (2010)
Yu, L.: A Developers Guide the Semantic Web. Springer (2011)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Geibel, P. et al. (2013). Ontology-Based Semantic Annotation of Documents in the Context of Patient Identification for Clinical Trials. In: Meersman, R., et al. On the Move to Meaningful Internet Systems: OTM 2013 Conferences. OTM 2013. Lecture Notes in Computer Science, vol 8185. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-41030-7_53
Download citation
DOI: https://doi.org/10.1007/978-3-642-41030-7_53
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-41029-1
Online ISBN: 978-3-642-41030-7
eBook Packages: Computer ScienceComputer Science (R0)