Zusammenfassung
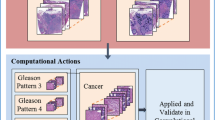
Prostate cancer (PCa) is the dominating malignant tumor for men worldwide and across all ethnic groups. If a carcinoma is being suspected, e.g. due to blood levels, trans-rectal punch biopsies of the prostate will be accomplished, while in case of higher stages of the disease the complete prostate is being surgically removed (radical prostatectomy). In both cases prostate tissue will be prepared into histological sections on glass microscope slides according to certain laboratory protocols, and is finally microscopically inspected by a trained histopathologist. Even though this method is well established, it can lead to various problems because of objectivity deficiencies. In this paper, we present a proof of concept of using Artificial Neural Networks (ANN) for automatically analyzing prostate cancer tissue and rating its malignancy using tissue microarrays (TMAs) of sampled benign and malignant tissue.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
Literatur
Gleason DF. Classification of prostatic carcinomas. Cancer Chemotherapy Reports. 1966;50(3):125–128.
Bostwick DG. Gleason grading of prostatic needle biopsies. correlation with grade in 316 matched prostatectomies. Am J Surg Pathol. 1994;18(8):796–803.
Allsbrook WC, Mangold KA, Johnson MH, et al. Interobserver reproducibility of gleason grading of prostatic carcinoma: urologic pathologists. Hum Pathol. 2001;32(1):74–80.
Doyle S, Feldman MD, Shih N, et al. Cascaded discrimination of normal, abnormal, and confounder classes in histopathology: gleason grading of prostate cancer. BMC Bioinformatics. 2012;13(282).
Sparks R, Madabhushi A. Statistical shape model for manifold regularization: gleason grading of prostate histology. Comput Vis Image Underst. 2013;117(9):1138–1146.
Loeer M, Greulich L, Scheibe P, et al. Classifying prostate cancer malignancy by quantitative histomorphometry. J Urol. 2012;187(5):1867–1875.
Salman S,Ma Z,Mohanty S, et al. A machine learning approach to identify prostate cancer areas in complex histological images. In: Pi, etka E, Kawa J, Wi,ec lawek W, editors. Information Technologies in Biomedicine, Volume 3. vol. 283 of Advances in Intelligent Systems and Computing. Springer International Publishing Switzerland; 2014. p. 295–306.
Arvaniti E, Fricker KS, Moret M, et al. Automated gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8(12054).
Howard AG, Zhu M, Chen B, et al. MobileNets: effcient convolutional neural networks for mobile vision applications; 2017. arXiv:1704.04861 [cs.CV].
Vahadane A, Peng T, Albarqouni S, et al. Structure-preserved color normalization for histological images. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI). IEEE; 2015. p. 1–4.
Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830.
Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving gleason scoring of prostate cancer. npj Digital Medicine. 2019;2(48).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Fachmedien Wiesbaden GmbH, ein Teil von Springer Nature
About this paper
Cite this paper
Bauer, M., Zürner, S., Popp, G., Kristiansen, G., Braumann, UD. (2020). Neural Network for Analyzing Prostate Cancer Tissue Microarrays. In: Tolxdorff, T., Deserno, T., Handels, H., Maier, A., Maier-Hein, K., Palm, C. (eds) Bildverarbeitung für die Medizin 2020. Informatik aktuell. Springer Vieweg, Wiesbaden. https://doi.org/10.1007/978-3-658-29267-6_4
Download citation
DOI: https://doi.org/10.1007/978-3-658-29267-6_4
Published:
Publisher Name: Springer Vieweg, Wiesbaden
Print ISBN: 978-3-658-29266-9
Online ISBN: 978-3-658-29267-6
eBook Packages: Computer Science and Engineering (German Language)