Abstract
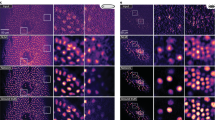
Examining specific sub-cellular structures while minimizing cell perturbation is important in the life sciences. Fluorescence labeling and imaging is widely used. With the advancement of deep learning, digital staining routines for label-free analysis have emerged to replace fluorescence imaging. Nonetheless, digital staining of sub-cellular structures such as mitochondria is sub-optimal. This is because the models designed for computer vision are directly applied instead of optimizing them for microscopy data. We propose a new loss function with multiple thresholding steps to promote more effective learning for microscopy data. We demonstrate a deep learning approach to translate the labelfree brightfield images of living cells into equivalent fluorescence images of mitochondria with an average structural similarity of 0.77, thus surpassing the state-of-the-art of 0.7 with L1. We provide insightful examples of unique opportunities by data-driven deep learning-enabled image translations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Samanta S, He Y, Sharma A, et al. Fluorescent probes for nanoscopic imaging of mitochondria. Chem. 2019;5(7):1697–1726.
Swayne TC, Gay AC, Pon LA. Methods cell biol.. vol. 80. Academic Press; 2007.
Chazotte B. Labelng mitochondria with fluorescent dyes for imaging. Cold Spring Harb Protoc. 2009;2009(6).
Kandel ME, Hu C, Kouzehgarani GN, et al. Epi-illumination gradient light interference microscopy for imaging opaque structures. Nat Commun. 2019;10(1):19.
Hoebe R, Van Oven C, Gadella TW, et al. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat Biotechnol. 2007;25(2):249–253.
Christiansen EM, Yang SJ, Ando DM, et al. In silico labeling: predicting fluorescent labels in unlabeled images. Cell. 2018;173(3):792–803.
Zahedi A, On V, Phandthong R, et al. Deep analysis of mitochondria and cell health using machine learning. Sci Rep. 2018;8(1):115.
Vicar T, Balvan J, Jaros J, et al. Cell segmentation methods for label-free contrast microscopy: review and comprehensive comparison. BMC Bioinformatics. 2019;20(1):360.
Mirza M, Osindero S. Conditional generative adversarial nets. arXiv preprint arXiv:14111784. 2014;.
Ronneberger O, Fischer P, Brox T; Springer. U-net: convolutional networks for biomedical image segmentation. CoRR. 2015; p. 234–241.
Isola P, Zhu JY, Zhou T, et al. Image-to-image translation with conditional adversarial networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2017; p. 5967–5976.
Armanious K, Jiang C, Fischer M, et al. MedGAN: medical image translation using GANs. Comput Med Imaging Graph. 2020;79:101684.
Kotte S, Kumar PR, Injeti SK. An efficient approach for optimal multilevel thresholding selection for gray scale images based on improved differential search algorithm. Ain Shams Med J. 2018;9(4):1043–1067.
Ounkomol C, Seshamani S, Maleckar MM, et al. Label-free prediction of threedimensional fluorescence images from transmitted-light microscopy. Nat Methods. 2018;15(11):917–920.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Der/die Autor(en), exklusiv lizenziert durch Springer Fachmedien Wiesbaden GmbH, ein Teil von Springer Nature
About this paper
Cite this paper
Somani, A. et al. (2021). Digital Staining of Mitochondria in Label-free Live-cell Microscopy. In: Palm, C., Deserno, T.M., Handels, H., Maier, A., Maier-Hein, K., Tolxdorff, T. (eds) Bildverarbeitung für die Medizin 2021. Informatik aktuell. Springer Vieweg, Wiesbaden. https://doi.org/10.1007/978-3-658-33198-6_55
Download citation
DOI: https://doi.org/10.1007/978-3-658-33198-6_55
Published:
Publisher Name: Springer Vieweg, Wiesbaden
Print ISBN: 978-3-658-33197-9
Online ISBN: 978-3-658-33198-6
eBook Packages: Computer Science and Engineering (German Language)