Abstract
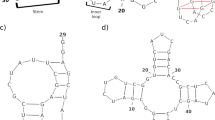
Nucleic acid nanotechnology offers many methods to fold into a variety of complex and functional nanostructures. These self-assemble scaffolds are valuable in various applications, such as molecular programming, sensing, drug delivery and nanofabrication. However, existing algorithms are typically lacking on predicting pseudoknots structure fast and accurately. This paper proposes a novel genetic algorithm to predict nucleic acid secondary structure including pseudoknots. The length of continues base pairs stacking and free energy are considered to evaluate individuals. Furthermore, the performance of our algorithm is compared with RNAStructure using PseudoBase benchmark instances, and the results show that our algorithm outperforms on accuracy and efficiency.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ishikawa, J., Furuta, H., Ikawa, Y.: RNA Tectonics (tectoRNA) for RNA nanostructure design and its application in synthetic biology. J. Wiley Interdisc. Rev. RNA 4, 651C–664 (2013)
Stewart, J.M., Franco, E.: Learning from DNA nanotechnology. DNA & RNA Nanotechnol. 2, 23–25 (2016)
Wang, C., Schröder, M.S., Hammel, S., Butler, G.: Using RNA-seq for analysis of differential gene expression in fungal species. In: Devaux, F. (ed.) Yeast Functional Genomics. MMB, vol. 1361, pp. 1–40. Springer, Heidelberg (2016). doi:10.1007/978-1-4939-3079-1_1
Marioni, J.C., Mason, C.E., Mane, S.M.: RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. J. Genome Res. 18, 1509–1517 (2008)
Andronescu, M., Bereg, V., Hoos, H.H., et al.: RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinform. 9, 1–10 (2008)
Zhang, X., Tian, Y., Cheng, R., Jin, Y.: An efficient approach to non-dominated sorting for evolutionary multi-objective optimiza-tion. IEEE Trans. Evol. Comput. 19, 201–213 (2015)
Anderson-Lee, J., Fisker, E., Kosaraju, V.: Principles for predicting RNA secondary structure design difficulty. J. Mol. Biol. 428, 748–757 (2016)
Wang, X., Song, T., Gong, F., Zheng, P.: On the computational power of spiking neural P systems with self-organization. Sci. Rep. doi:10.1038/srep27624
Leonard, C.W., Weeks, K.M.: RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA Publ. RNA Soc. 20, 846–854 (2014)
Waterman, M.S., Smith, T.F.: Rapid dynamic programming algorithms for RNA secondary structure. Adv. Appl. Math. 7, 455–464 (1986)
Can, D., Narayanan, S.: A dynamic programming algorithm for computing N-gram posteriors from lattices. In: Conference on Empirical Methods in Natural Language Processing (2015)
Song, T., Zeng, X., Liu, X.: Asynchronous spiking neural P systems with rules on synapses. Neurocomputing 151, 1439–1445 (2015)
Xingyi, Z., Linqiang, P., Andrei, P.: On universality of axon P systems. IEEE Trans. Neural Netw. Learn. Syst. 26, 2816–2829 (2015)
Pan, L., Wang, J., Hoogeboom, H.J.: Spiking neural P systems with astrocytes. Neural Comput. 24, 805–825 (2012)
Song, T., Zheng, P., Wong, M.D., Wang, X.: Design of logic gates using spiking neural P systems with homogeneous neurons and astrocytes-like control. Inf. Sci. 372, 380–391 (2016)
Zeng, X., Zhang, X., Song, T., Pan, L.: Spiking neural P systems with thresholds. Neural Comput. 26, 1340–1361 (2014)
Xu, J.: Probe machine. IEEE Trans. Neural Netw. Learn. Syst. 27, 1405–1416 (2016)
Nawrocki, E.P., Burge, S.W., Bateman, A., et al.: Rfam 12.0: updates to the RNA families database. J. Nucleic Acids Res. 43, 130-7 (2015)
Shi, X., Wu, X., Song, T., Li, X.: Construction of DNA nanotubeswith controllable diameters and patterns by using hierarchical DNA sub-tiles. Nanoscale. doi:10.1039/C6NR02695H
Shi, X., Wang, Z., Deng, C., Song, T., Pan, L., Chen, Z.: A novel bio-sensor based on DNA strand displacement, PloS ONE 9(10), e108856
Shi, X., Chen, C., Li, X., Song, T., Chen, Z., Zhang, Z., Wang, Y.: Size controllable DNA nanoribbons assembled from three types of reusable brick single-strand DNA tiles. 11(43), 8484–8492 (2015)
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant Nos. 61472293, 61502012, 60974112 and 91130034), Natural Science Foundation of Hubei Province (2015CFB335), and the Beijing Natural Science Foundation (4164096).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Kai, Z., Shangyi, L., Juanjuan, H., Yunyun, N. (2016). Nucleic Acid Secondary Structures Prediction with Planar Pseudoknots Using Genetic Algorithm. In: Gong, M., Pan, L., Song, T., Zhang, G. (eds) Bio-inspired Computing – Theories and Applications. BIC-TA 2016. Communications in Computer and Information Science, vol 682. Springer, Singapore. https://doi.org/10.1007/978-981-10-3614-9_54
Download citation
DOI: https://doi.org/10.1007/978-981-10-3614-9_54
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-3613-2
Online ISBN: 978-981-10-3614-9
eBook Packages: Computer ScienceComputer Science (R0)