Abstract
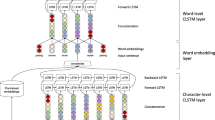
Performance enhancement of biomedical named entity recognition tagging by applying a deep learning based framework is introduced with a combination of the word as well as character embedding. The input sentences first pass through the word and character level embedding, where word embedding is used to learn syntactic and semantic information and character level embedding handles those words which are out-of-vocabulary, then it passes through Bi-directional Long Short-Term Memory (BI-LSTM), where first it train the sentences in the forward direction and then again it trains the sentences in the backward direction, and finally it passes through CRF layer where the output comes in the form of gene mention tagging. The framework is tested over BioCreative II Gene mention task corpus. The deep learning framework combines with the conditional random field, and embedding techniques achieve 89.42% of F-score, which outperforms the various state-of-the-art techniques and top-ranked achieved system on BioCreative II Gene Mention competition.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chang, Y.M., Kuo, C.J., Huang, H.S., Lin, Y.S., Hsu, C.N.: Analysis and enhancement of conditional random fields gene mention taggers in BioCreative II challenge evaluation. LBM (Short Papers) 7(1) (2007)
Wang, L., Sun, Y., Zhu, Z.: Knowledge points extraction of junior high school english exercises based on SVM method. In: Proceedings of the 2nd International Conference on E-Education, E-Business and E-Technology, pp. 43–47. ACM, New York, United State (2018)
Smith, L., Tanabe, L.K., nee Ando, R.J., Kuo, C.J., Chung, I.F., Hsu, C.N., Torii, M.: Overview of BioCreative II gene mention recognition. Genome Biol. 9(2), S2 (2008)
Li, L., Sun, J., Huang, D.: Boosting performance of gene mention tagging system by classifiers ensemble. In: Proceedings of the 6th International Conference on Natural Language Processing and Knowledge Engineering (NLPKE-2010), pp. 1–4. IEEE, Beijing, China (2010)
Ando, R.K.: BioCreative II gene mention tagging system at IBM Wat-son. In: Proceedings of the Second BioCreative Challenge Evaluation Workshop, pp. 101–103. Centro Nacional de Investigaciones Oncologicas (CNIO) Madrid, Spain (2007)
Kuo, C.J., Chang, Y.M., Huang, H.S., Lin, K.T., Yang, B.H., Lin, Y.S., Chung, I.F.: Rich feature set, unification of bidirectional parsing and dictionary filtering for high F-score gene mention tagging. In: Proceedings of the Second BioCreative Challenge Evaluation Workshop, pp. 105–107. Centro Nacional de Investigaciones Oncologicas (CNIO) Madrid, Spain (2007)
Yang, X., Gao, Z., Li, Y., Pan, C., Yang, R., Gong, L., Yang, G.: Bidirectional LSTM-CRF for biomedical named entity recognition. In: 14th International Conference on Natural Computation, Fuzzy Systems and Knowledge Discovery (ICNC-FSKD), pp. 239–242. IEEE, Xi’an, China (2018)
Sutton, C., McCallum, A.: An Introduction to Conditional Random Fields. arXiv, p. arXiv preprint arXiv:1011.4088 (2010)
Laddha, A., Mukherjee, A.: Aspect Specific Opinion Expression Extraction using Attention based LSTM-CRF Network (2019). arXiv preprint arXiv:1902.02709
BioCreative Homepage. https://biocreative.bioinformatics.udel.edu. Last ccessed 01 Nov 2019
Huang, M.S., Lai, P.T., Tsai, R.T.H., Hsu, W. L.: Revised JNLPBA Corpus: A Revised Version of Biomedical NER Corpus for Relation Extraction Task (2019). arXiv preprint arXiv:1901.10219 (2019)
Acknowledgements
The authors would like to thank the National Institute of Technology Raipur, India for the motivation and support for doing this research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Kumar, A., Sharaff, A. (2021). Performance Enhancement of Gene Mention Tagging by Using Deep Learning and Biomedical Named Entity Recognition. In: Satapathy, S., Zhang, YD., Bhateja, V., Majhi, R. (eds) Intelligent Data Engineering and Analytics. Advances in Intelligent Systems and Computing, vol 1177. Springer, Singapore. https://doi.org/10.1007/978-981-15-5679-1_61
Download citation
DOI: https://doi.org/10.1007/978-981-15-5679-1_61
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-5678-4
Online ISBN: 978-981-15-5679-1
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)