Abstract
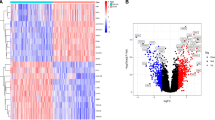
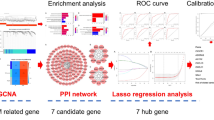
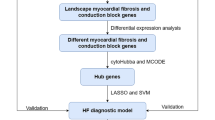
Objective: Screening for dilated cardiomyopathy (DCM) core genes and letter immune infiltration by bioinformatic methods to find new strategies for prevention and treatment. Methods: Gene expression compilation database (GEO) GSE3585 and GSE17800 gene microarray sets were extracted and differentially expressed genes (DEGs) were obtained from DCM and normal control myocardial biopsies using R language. The DEGs were tested for gene ontology (GO) functional analysis, Kyoto Gene and Genome Encyclopedia (KEGG) pathway analysis and gene probe (GSEA) enrichment. The Lasso algorithm was subsequently used to identify key DCM-related genes in the practice set and to authenticate them against the test set. Prospective mechanisms for DCM development included: differences in key gene expression between normal and DCM samples, variations in western blot coverage, correlates of clinical relevance to exempt cells, and key gene and immune cell correlation. Results: The final screening identified 2 key genes, NPPA and NPPB. The variation in expression of the key genes between normal and DCM samples can be regarded as a diagnostic factor for patients. Also, There is a striking divergence in vaccine levels of immuinia among normal and DCM samples, and the critical gene expression was closely related to the richness of immune cell infiltration. Conclusion: Based on bioinformatics analysis and review of relevant literature, two candidate genes, NPPA and NPPB, were screened and strongly associated with the progression of DCM, providing meaningful clues and suggestions used to prevent and heal DCM.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Merlo M, Cannatà A, Gobbo M, Stolfo D, Elliott PM, Sinagra G. Evolving concepts in dilated cardiomyopathy. Eur J Heart Fail. 2018;20(2):228–239. https://doi.org/10.1002/ejhf.1103.
Priori SG, Blomström-Lundqvist C, Mazzanti A, et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC). Eur Heart J. 2015;36(41):2793–2867. https://doi.org/10.1093/eurheartj/ehv316
Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(D1):D991–5.
Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One. 2013;8(3):e57886. https://doi.org/10.1371/journal.pone.0057886.
Deng YJ, Ren EH, Yuan WH, Zhang GZ, Wu ZL, Xie QQ. GRB10 and E2F3 as Diagnostic Markers of Osteoarthritis and Their Correlation with Immune Infiltration. Diagnostics (Basel). 2020;10(3):171. Published 2020 Mar 22. https://doi.org/10.3390/diagnostics10030171
Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. https://doi.org/10.1093/nar/gkv007
Weintraub RG, Semsarian C, Macdonald P. Dilated cardiomyopathy. Lancet. 2017;390(10092):400–414. https://doi.org/10.1016/S0140-6736(16)31713-5
Yu G, Wang LG, Han Y, et al. clusterProfiler: An R package for comparing biological themes among gene clusters [J]. OMICS, 2012, 16(5): 284–287.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. https://doi.org/10.1089/omi.2011.0118.
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. https://doi.org/10.1038/75556.
Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. Published 2011 Mar 17. https://doi.org/10.1186/1471-2105-12-77
Liu, S., Xiao, Z., You, X. and Su, R., 2022. Multistrategy boosted multicolony whale virtual parallel optimization approaches. Knowledge-Based Systems, 242, p. 108341.
Huang, W., Li, Y., Zhang, K., Hou, X., Xu, J., Su, R. and Xu, H., 2021. An Efficient Multi-Scale Focusing Attention Network for Person Re-Identification. Applied Sciences, 11(5), p. 2010.
Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. Published 2011 Mar 17. https://doi.org/10.1186/1471-2105-12-77.
Yang L, Shou YH, Yang YS, Xu JH. Elucidating the immune infiltration in acne and its comparison with rosacea by integrated bioinformatics analysis. PLoS One. 2021;16(3):e0248650. Published 2021 Mar 24. https://doi.org/10.1371/journal.pone.0248650.
Cao Y, Tang W, Tang W. Immune cell infiltration characteristics and related core genes in lupus nephritis: results from bioinformatic analysis. BMC Immunol. 2019;20(1):37. Published 2019 Oct 21. https://doi.org/10.1186/s12865-019-0316-x
Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. https://doi.org/10.1093/bioinformatics/btv300
Ware JS, Cook SA. Role of titin in cardiomyopathy: from DNA variants to patient stratification. Nat Rev Cardiol. 2018;15(4):241–252. https://doi.org/10.1038/nrcardio.2017.190
Mazzarotto F, Tayal U, Buchan RJ, et al. Reevaluating the Genetic Contribution of Monogenic Dilated Cardiomyopathy. Circulation. 2020;141(5):387–398. https://doi.org/10.1161/CIRCULATIONAHA.119.037661
Wood Heickman LK, DeBoer MD, Fasano A. Zonulin as a potential putative biomarker of risk for shared type 1 diabetes and celiac disease autoimmunity. Diabetes Metab Res Rev. 2020;36(5):e3309. https://doi.org/10.1002/dmrr.3309
Asbjornsdottir B, Snorradottir H, Andresdottir E, et al. Zonulin-Dependent Intestinal Permeability in Children Diagnosed with Mental Disorders: A Systematic Review and Meta-Analysis. Nutrients. 2020;12(7):1982. Published 2020 Jul 3. https://doi.org/10.3390/nu12071982
Iwata Y, Ito S, Wakabayashi S, Kitakaze M. TRPV2 channel as a possible drug target for the treatment of heart failure. Lab Invest. 2020;100(2):207–217. https://doi.org/10.1038/s41374-019-0349-z
Zhu YX, Huang JQ, Ming YY, Zhuang Z, Xia H. Screening of key biomarkers of tendinopathy based on bioinformatics and machine learning algorithms. PLoS One. 2021;16(10):e0259475. Published 2021 Oct 29. https://doi.org/10.1371/journal.pone.0259475
Acknowledgements
This study was funded by the S&T Innovation Project for Universities in Shanxi Province (2019L0683), the Graduate Education Innovation Project in Shanxi Province (2022Y37) and the Provincial Science and Technology Grant in Shanxi Province (20210302124588).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Bi, X., Zhan, Z., Yang, J., Tang, X., Zhao, T. (2023). Machine Learning for the Evaluation and Detection of Key Markers in Dilated Cardiomyopathy. In: Su, R., Zhang, Y., Liu, H., F Frangi, A. (eds) Medical Imaging and Computer-Aided Diagnosis. MICAD 2022. Lecture Notes in Electrical Engineering, vol 810. Springer, Singapore. https://doi.org/10.1007/978-981-16-6775-6_42
Download citation
DOI: https://doi.org/10.1007/978-981-16-6775-6_42
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-6774-9
Online ISBN: 978-981-16-6775-6
eBook Packages: MedicineMedicine (R0)