Abstract
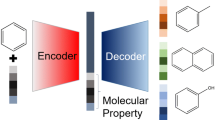
In recent decades, there has been a significant increase in the application of deep learning in drug design. We present a basic method for generating diverse molecules using variational autoencoder. The variational autoencoder is based on recurrent neural network. A string representation SMILES of molecules is used for training the proposed model. A parameter that can be tweaked determines the level of diversity. Interpolation is also performed between two drug molecules in the latent space. The diverse variational autoencoder (dVAE) shows superior results as compare with other state-of-the-art methods. Thus, it can be used to generate controllable diverse molecules.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Xu, Y., et al.: Deep learning for molecular generation. Future Med. Chem. 11(6), 567–597 (2019)
Elton, D.C., et al.: Deep learning for molecular design-a review of the state of the art. Mol. Syst. Des. Eng. 4(4), 828–849 (2019)
Vamathevan, J., et al.: Applications of machine learning in drug discovery and development. Nature Rev. Drug Discovery 18(6), 463–477 (2019)
Sanchez-Lengeling, Benjamin, Aspuru-Guzik, Aláin.: Inverse molecular design using machine learning: generative models for matter engineering. Science 361(6400), 360–365 (2018)
Lopyrev, K.: Generating news headlines with recurrent neural networks. arXiv preprint arXiv:1512.01712 (2015)
Briot, J.-P., Hadjeres, G., Pachet, F.-D.: Deep learning techniques for music generation. Springer (2020)
Wang, Z., He, W., Wu, H., Wu, H., Li, W., Wang, H., Chen, E.E.: Chinese poetry generation with planning based neural network. arXiv preprint arXiv:1610.09889 (2016)
Elgammal, A., et al.: Can: creative adversarial networks, generating art by learning about styles and deviating from style norms. arXiv preprint arXiv:1706.07068 (2017)
Segler, M.H., Preuss, M., Waller, M.P.: Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555(7698), 604–610 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241. Springer
Chen, H., et al.: The rise of deep learning in drug discovery. Drug Discovery Today 23(6), 1241–1250 (2018)
Weininger, D: SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28(1), 31–36 (1988)
Schwalbe-Koda, D., Gómez-Bombarelli, R.: Generative models for automatic chemical design. In: Machine Learning Meets Quantum Physics, pp. 445–467. Springer, Cham (2020)
Arús-Pous, J., et al.: Exploring the GDB-13 chemical space using deep generative models. J. Cheminformatics 11(1), 1–14 (2019)
Polykovskiy, D., et al.: Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol 11, 1931 (2020)
Gómez-Bombarelli, R., et al.: Automatic chemical design using a data-driven continuous representation of molecules. ACS Central Sci. 4(2), 268-276 (2018)
Winter, R., et al.: Efficient multi-objective molecular optimization in a continuous latent space. Chem. Sci. 10(34), 8016–8024 (2019)
Jannik Bjerrum, E., Sattarov, B.: Improving chemical autoencoder latent space and molecular De novo generation diversity with heteroencoders. arXiv e-prints: arXiv-1806 (2018)
Lim, J., et al.: Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminformatics 10(1), 1–9 (2018)
Landrum, G.: RDKit: Open-source cheminformatics. (Online). http://wwwrdkit.org. Accessed 3 Jan 2022, 2012 (2006)
Kingma, D.P., Welling, M.: Auto-encoding variational bayes. arXiv preprint arXiv:1312.6114 (2013)
Sutskever, I., Vinyals, O., Le, Q.V.: Sequence to sequence learning with neural networks. Adv. Neural Inf. Process. Syst. 27 (2014)
Preuer, K., et al.: Fréchet ChemNet distance: a metric for generative models for molecules in drug discovery. J. Chem. Inf. Model. 58(9), 1736–1741 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Singh Bhadwal, A., Kumar, K. (2023). Direct De Novo Molecule Generation Using Probabilistic Diverse Variational Autoencoder. In: Tistarelli, M., Dubey, S.R., Singh, S.K., Jiang, X. (eds) Computer Vision and Machine Intelligence. Lecture Notes in Networks and Systems, vol 586. Springer, Singapore. https://doi.org/10.1007/978-981-19-7867-8_2
Download citation
DOI: https://doi.org/10.1007/978-981-19-7867-8_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-7866-1
Online ISBN: 978-981-19-7867-8
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)