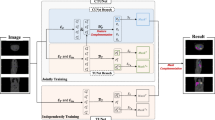
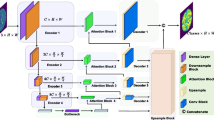
Abstract
Deep learning has been extensively employed in the field of medical image segmentation, demonstrating its robustness and efficacy. However, the pursuit of consistent segmentation performance across diverse instrumental conditions and the challenge of achieving precise boundary delineation in segmented images remain significant hurdles. In this paper, we aim to develop a model capable of achieving consistent, high-quality segmentation of identical regions of interest across varying instrumental conditions, with precise boundary delineation. Toward this end, we introduce our Hybrid Dynamic MedNeXt (HDNeXt) model, an advanced framework capable of dynamically generating weights across diverse medical images to maintain consistently high segmentation performance. HDNeXt builds on the robust segmentation framework of MedNeXt by incorporating dynamic convolution techniques, which endow the model with the capability for dynamic weight adjustment, significantly enhancing its segmentation performance. To tackle the second challenge, we devised a novel loss function, \(L_{CR}\), formulated on the Curvature of the segmentation boundary and Region-Fitting energy derived from level set methods, which significantly enhances boundary precision during training and optimizes overall segmentation performance. Experiments were conducted on the abdominal CT datasets Synapse and the cardiac MRI datasets ACDC to demonstrate the efficiency and effectiveness of our method. Our method achieved an average Dice coefficient of 84.38 on the Synapse datasets and 93.59 on the ACDC datasets, surpassing other 2D state-of-the-art segmentation models and achieving optimal performance for 2D medical image segmentation. Codes are available at https://github.com/HaoyuCao/HDNeXt.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., Wang, M.: Swin-unet: Unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision. pp. 205–218. Springer (2022)
Chambolle, A., Pock, T.: Total roto-translational variation. Numer. Math. 142, 611–666 (2019)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–277 (2001)
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y.: Transunet: Transformers make strong encoders for medical image segmentation. ArXiv preprint arXiv:2102.04306 (2021)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2017)
Chen, Y., Dai, X., Liu, M., Chen, D., Yuan, L., Liu, Z.: Dynamic convolution: Attention over convolution kernels. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 11030–11039 (2020)
Dai, Z., Liu, H., Le, Q.V., Tan, M.: Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural. Inf. Process. Syst. 34, 3965–3977 (2021)
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. ArXiv preprint arXiv:2010.11929 (2020)
Ha, D.T., Phuong, D.L.: Freedom of information law comes to vietnam: How do human rights adapt to goals of economic development and political stability? Austl. J. Asian L. 18, 167 (2017)
Han, Q., Fan, Z., Dai, Q., Sun, L., Cheng, M.M., Liu, J., Wang, J.: On the connection between local attention and dynamic depth-wise convolution. ArXiv preprint arXiv:2106.04263 (2021)
Huang, T., Huang, L., You, S., Wang, F., Qian, C., Xu, C.: Lightvit: Towards light-weight convolution-free vision transformers. arXiv preprint arXiv:2207.05557 (2022)
Huang, X., Deng, Z., Li, D., Yuan, X.: Missformer: An effective medical image segmentation transformer. arXiv preprint arXiv:2109.07162 (2021)
Huang, Z., Zhang, Z., Lan, C., Zha, Z.J., Lu, Y., Guo, B.: Adaptive frequency filters as efficient global token mixers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 6049–6059 (2023)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Kim, B., Ye, J.C.: Mumford-shah loss functional for image segmentation with deep learning. IEEE Trans. Image Process. 29, 1856–1866 (2019)
Kim, Y., Kim, S., Kim, T., Kim, C.: Cnn-based semantic segmentation using level set loss. In: 2019 IEEE winter conference on applications of computer vision (WACV). pp. 1752–1760. IEEE (2019)
Kuiper, N.H.: Minimal total absolute curvature for immersions. Invent. Math. 10(3), 209–238 (1970)
Langer, J., Singer, D.A.: The total squared curvature of closed curves. Journal of Differential Geometry 20(1), 1–22 (1984)
Li, C., Zhou, A., Yao, A.: Omni-dimensional dynamic convolution. ArXiv preprint arXiv:2209.07947 (2022)
Li, C., Gore, J.C., Davatzikos, C.: Multiplicative intrinsic component optimization (mico) for mri bias field estimation and tissue segmentation. Magn. Reson. Imaging 32(7), 913–923 (2014)
Li, C., Kao, C.Y., Gore, J.C., Ding, Z.: Minimization of region-scalable fitting energy for image segmentation. IEEE Trans. Image Process. 17(10), 1940–1949 (2008)
Liu, C., Chen, L.C., Schroff, F., Adam, H., Hua, W., Yuille, A.L., Fei-Fei, L.: Auto-deeplab: Hierarchical neural architecture search for semantic image segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 82–92 (2019)
Liu, Z., Hu, H., Lin, Y., Yao, Z., Xie, Z., Wei, Y., Ning, J., Cao, Y., Zhang, Z., Dong, L., et al.: Swin transformer v2: Scaling up capacity and resolution. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 12009–12019 (2022)
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin, S., Guo, B.: Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 10012–10022 (2021)
Liu, Z., Mao, H., Wu, C.Y., Feichtenhofer, C., Darrell, T., Xie, S.: A convnet for the 2020s. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 11976–11986 (2022)
Ma, D., Liao, Q., Chen, Z., Liao, R., Ma, H.: Adaptive local-fitting-based active contour model for medical image segmentation. Signal Processing: Image Communication 76, 201–213 (2019)
Niu, S., Chen, Q., De Sisternes, L., Ji, Z., Zhou, Z., Rubin, D.L.: Robust noise region-based active contour model via local similarity factor for image segmentation. Pattern Recogn. 61, 104–119 (2017)
Peng, C., Zhang, X., Yu, G., Luo, G., Sun, J.: Large kernel matters–improve semantic segmentation by global convolutional network. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 4353–4361 (2017)
Rahman, M.M., Marculescu, R.: Medical image segmentation via cascaded attention decoding. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. pp. 6222–6231 (2023)
Rahman, M.M., Marculescu, R.: G-cascade: Efficient cascaded graph convolutional decoding for 2d medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. pp. 7728–7737 (2024)
Rahman, M.M., Marculescu, R.: Multi-scale hierarchical vision transformer with cascaded attention decoding for medical image segmentation. In: Medical Imaging with Deep Learning. pp. 1526–1544. PMLR (2024)
Ren, M., Triantafillou, E., Ravi, S., Snell, J., Swersky, K., Tenenbaum, J.B., Larochelle, H., Zemel, R.S.: Meta-learning for semi-supervised few-shot classification. ArXiv preprint arXiv:1803.00676 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18. pp. 234–241. Springer (2015)
Roy, S., Koehler, G., Ulrich, C., Baumgartner, M., Petersen, J., Isensee, F., Jaeger, P.F., Maier-Hein, K.H.: Mednext: transformer-driven scaling of convnets for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 405–415. Springer (2023)
Sun, C., Shrivastava, A., Singh, S., Gupta, A.: Revisiting unreasonable effectiveness of data in deep learning era. In: Proceedings of the IEEE International Conference on Computer Vision. pp. 843–852 (2017)
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I.: Attention is all you need. Advances in Neural Information Processing Systems 30 (2017)
Woo, S., Debnath, S., Hu, R., Chen, X., Liu, Z., Kweon, I.S., Xie, S.: Convnext v2: Co-designing and scaling convnets with masked autoencoders. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 16133–16142 (2023)
Wu, J., Ji, W., Fu, H., Xu, M., Jin, Y., Xu, Y.: Medsegdiff-v2: Diffusion-based medical image segmentation with transformer. In: Proceedings of the AAAI Conference on Artificial Intelligence. vol. 38, pp. 6030–6038 (2024)
Xie, Z., Zhang, Z., Cao, Y., Lin, Y., Bao, J., Yao, Z., Dai, Q., Hu, H.: Simmim: A simple framework for masked image modeling. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 9653–9663 (2022)
Xu, K., Qin, M., Sun, F., Wang, Y., Chen, Y.K., Ren, F.: Learning in the frequency domain. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 1740–1749 (2020)
Yang, B., Bender, G., Le, Q.V., Ngiam, J.: Condconv: Conditionally parameterized convolutions for efficient inference. Advances in Neural Information Processing Systems 32 (2019)
Yang, Y., Yan, T., Jiang, X., Xie, R., Li, C., Zhou, T.: Mh-net: Model-data-driven hybrid-fusion network for medical image segmentation. Knowl.-Based Syst. 248, 108795 (2022)
Yu, W., Zhou, P., Yan, S., Wang, X.: Inceptionnext: When inception meets convnext. arXiv preprint arXiv:2303.16900 (2023)
Zhou, H.Y., Guo, J., Zhang, Y., Yu, L., Wang, L., Yu, Y.: nnformer: Interleaved transformer for volumetric segmentation. ArXiv preprint arXiv:2109.03201 (2021)
Zhou, Z., Siddiquee, M.M.R., Tajbakhsh, N., Liang, J.: Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 39(6), 1856–1867 (2019)
Acknowledgments
This research is supported by National Natural Science Foundation of China No. 62371156 and Natural Science Foundation of Guangdong Province No. 2022A1515011629, University Innovative Team Project of Guangdong 2022KCXTD0 39, and National Natural Science Foundation of China (12371419).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2025 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Cao, H., Han, T., Yang, Y. (2025). HDNeXt: Hybrid Dynamic MedNeXt with Level Set Regularization for Medical Image Segmentation. In: Cho, M., Laptev, I., Tran, D., Yao, A., Zha, H. (eds) Computer Vision – ACCV 2024. ACCV 2024. Lecture Notes in Computer Science, vol 15478. Springer, Singapore. https://doi.org/10.1007/978-981-96-0963-5_24
Download citation
DOI: https://doi.org/10.1007/978-981-96-0963-5_24
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-96-0962-8
Online ISBN: 978-981-96-0963-5
eBook Packages: Computer ScienceComputer Science (R0)