Abstract
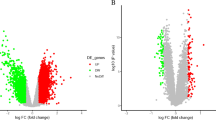
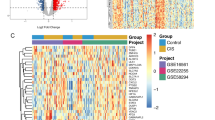
Ischemic Stroke (IS) has been ever-threatening to public health. Oxidative stress participated in the IS process and induced neuron death, lacking pathological elucidation and clinical diagnosis. Acquiring data from Gene Expression Omnibus (GEO) and Gene Set Enrichment Analysis (GSEA) database, we screened differently expressed genes and selected candidate diagnosis-related genes via random forest and SVM-RFE method. Subsequently we established a corresponding diagnosis model and eventually conducted the immune infiltration analysis. Parallelly, we conducted the consensus clustering and performed weighted gene co-expression network analysis (WGCNA) to figure out co-expressed gene modules in clusters respectively. Then we conducted functional enrichment for featured pathways of each cluster, and presented the KEGG pathways on the proteomic level. Eventually, the single-cell analysis was conducted to further explore the oxidative stress in IS, including pseudo-time analysis and cell-cell communication. Through machine learning algorithms, 9 key hub genes were selected and a diagnosis prediction model was conducted. The immune infiltration analysis revealed correlations between immune cells, inflammatory factors and differently expressed genes. The clusters obtained by consensus clustering indicated different functional pathways. The single-cell analysis revealed that endothelial cells were obviously affected by oxidative stress in IS. Associations between oxidative stress-induced and IS was clarified. New perspectives were provided for IS pathological elucidation, diagnosis and treatment usage.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lallukka, T., et al.: Trends in diagnosis-specific work disability before and after stroke: a longitudinal population-based study in Sweden. J. Am. Heart Assoc. 7(1), e006991 (2018). https://doi.org/10.1161/JAHA.117.006991
Virtanen, M., et al.: Work disability before and after a major cardiovascular event: a ten-year study using nationwide medical and insurance registers. Sci. Rep. 7(1), 1142 (2017). https://doi.org/10.1038/s41598-017-01216-2
Mendelson, S.J., Prabhakaran, S.: Diagnosis and management of transient ischemic attack and acute ischemic stroke: a review. JAMA. 325(11), 1088–1098 (2021). https://doi.org/10.1001/jama.2020.26867
Rammal, S.A., Almekhlafi, M.A.: Diabetes mellitus and stroke in the Arab world. J. Taibah Univ. Med. Sci. 11(4), 295–300 (2016)
Campbell, B.C.V., Khatri, P.: Stroke. Lancet 396(10244), 129–142 (2020). https://doi.org/10.1016/S0140-6736(20)31179-X
Sommer, C.J.: Ischemic stroke: experimental models and reality. Acta Neuropathol. 133(2), 245–261 (2017). https://doi.org/10.1007/s00401-017-1667-0
He, Z., Ning, N., Zhou, Q., Khoshnam, S.E., Farzaneh, M.: Mitochondria as a therapeutic target for ischemic stroke. Free Radic. Biol. Med. 146, 45–58 (2020). https://doi.org/10.1016/j.freeradbiomed.2019.11.005. Epub 5 November 2019
Jayaraj, R.L., Azimullah, S., Beiram, R., Jalal, F.Y., Rosenberg, G.A.: Neuroinflammation: friend and foe for ischemic stroke. J. Neuroinflamm. 16(1), 142 (2019). https://doi.org/10.1186/s12974-019-1516-2
Wu, S., Cheng, Y., Wu, B., Liu, M.: Stroke research in 2019: towards optimising treatment and prevention. Lancet Neurol. 19(1), 2–3 (2020). https://doi.org/10.1016/S1474-4422(19)30448-X
Rodrigues, F.B., Neves, J.B., Caldeira, D., Ferro, J.M., Ferreira, J.J., Costa, J.: Endovascular treatment versus medical care alone for ischaemic stroke: systematic review and meta-analysis. BMJ. 353, i1754 (2016). https://doi.org/10.1136/bmj.i1754
Li, Z., et al.: The role of oxidative stress in acute ischemic stroke-related thrombosis. Oxid. Med. Cell. Longev. 2022, 8418820 (2022). https://doi.org/10.1155/2022/8418820
Mathews, M.T., Berk, B.C.: PARP-1 inhibition prevents oxidative and nitrosative stress-induced endothelial cell death via transactivation of the VEGF receptor 2. Arterioscler. Thromb. Vasc. Biol. 28(4), 711–717 (2008)
Li, Y., et al.: Febuxostat attenuates paroxysmal atrial fibrillation-induced regional endothelial dysfunction. Thromb. Res. 149, 17–24 (2017). https://doi.org/10.1016/j.thromres.2016.11.011
Vexler, Z.S., Yenari, M.A.: Does inflammation after stroke affect the developing brain differently than adult brain? Dev. Neurosci. 31(5), 378–393 (2009). https://doi.org/10.1159/000232556
Fernandes, E.F.A., Özcelik, D.: Imaging biomarkers for monitoring the inflammatory redox landscape in the brain. Antioxidants 10(4), 528 (2021). https://doi.org/10.3390/antiox10040528
O’Connell, G.C., et al.: Machine-learning approach identifies a pattern of gene expression in peripheral blood that can accurately detect ischaemic stroke. NPJ Genomic Med. 1, 16038 (2016). https://doi.org/10.1038/npjgenmed.2016.38
Yu, H., et al.: Integrated transcriptomics reveals the brain and blood biomarkers in Alzheimer’s disease. CNS Neurosci. Ther. (2023). https://doi.org/10.1111/cns.14316
ADVANCE Collaborative Group, et al.: Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N. Engl. J. Med. 358(24), 2560–2572 (2008). https://doi.org/10.1056/NEJMoa0802987
Wang, T.J., et al.: Multiple biomarkers for the prediction of first major cardiovascular events and death. N. Engl. J. Med. 355(25), 2631–2639 (2006). https://doi.org/10.1056/NEJMoa055373
Stamova, B., et al.: Gene expression in peripheral immune cells following cardioembolic stroke is sexually dimorphic. PLoS One. 9(7), e102550 (2014). https://doi.org/10.1371/journal.pone.0102550
Zheng, K., et al.: Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J. Cereb. Blood Flow Metab. 42(1), 56–73 (2022). https://doi.org/10.1177/0271678X211026770
Subramanian, A., et al.: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102(43), 15545–15550 (2005). https://doi.org/10.1073/pnas.0506580102
Irizarry, R.A., et al.: Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4(2), 249–264 (2003). https://doi.org/10.1093/biostatistics/4.2.249
Buus, T.B., Ødum, N., Geisler, C., Lauritsen, J.P.H.: Three distinct developmental pathways for adaptive and two IFN-γ-producing γδ T subsets in adult thymus. Nat. Commun. 8(1), 1911 (2017). https://doi.org/10.1038/s41467-017-01963-w
Hoang, S.A., et al.: Gene expression predicts histological severity and reveals distinct molecular profiles of nonalcoholic fatty liver disease. Sci. Rep. 9(1), 12541 (2019). https://doi.org/10.1038/s41598-019-48746-5
Ritchie, M.E., et al.: limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47 (2015). https://doi.org/10.1093/nar/gkv007. Epub 20 January 2015
Sanz, H., Valim, C., Vegas, E., Oller, J.M., Reverter, F.: SVM-RFE: selection and visualization of the most relevant features through non-linear kernels. BMC Bioinform. 19(1), 432 (2018). https://doi.org/10.1186/s12859-018-2451-4
Zeng, D., et al.: IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol. 12, 687975 (2021). https://doi.org/10.3389/fimmu.2021.687975
Newman, A.M., et al.: Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12(5), 453–457 (2015). https://doi.org/10.1038/nmeth.3337
Becht, E., et al.: Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17(1), 218 (2016). https://doi.org/10.1186/s13059-016-1070-5
Wilkerson, M.D., Hayes, D.N.: ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26(12), 1572–1573 (2010). https://doi.org/10.1093/bioinformatics/btq170
Langfelder, P., Horvath, S.: WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008). https://doi.org/10.1186/1471-2105-9-559
Liebermeister, W., Noor, E., Flamholz, A., Davidi, D., Bernhardt, J., Milo, R.: Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. U S A 111(23), 8488–8493 (2014). https://doi.org/10.1073/pnas.1314810111
Jin, S., et al.: Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12(1), 1088 (2021). https://doi.org/10.1038/s41467-021-21246-9
Castroflorio, E., et al.: The Ncoa7 locus regulates V-ATPase formation and function, neurodevelopment and behaviour. Cell. Mol. Life Sci. 78(7), 3503–3524 (2021). https://doi.org/10.1007/s00018-020-03721-6
Chen, D., Fan, Z., Rauh, M., Buchfelder, M., Eyupoglu, I.Y., Savaskan, N.: ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene 36(40), 5593–5608 (2017). https://doi.org/10.1038/onc.2017.146
Li, W., et al.: Nono deficiency compromises TET1 chromatin association and impedes neuronal differentiation of mouse embryonic stem cells. Nucleic Acids Res. 48(9), 4827–4838 (2020). https://doi.org/10.1093/nar/gkaa213
Tavakoli Dargani, Z., Singla, D.K.: Embryonic stem cell-derived exosomes inhibit doxorubicin-induced TLR4-NLRP3-mediated cell death-pyroptosis. Am. J. Physiol. Heart Circ. Physiol. 317(2), H460–H471 (2019). https://doi.org/10.1152/ajpheart.00056.2019
Qiao, J, et al.: Targeting tumors with IL-10 prevents dendritic cell-mediated CD8+ T cell apoptosis. Cancer Cell 35(6), 901–915.e4 (2019). https://doi.org/10.1016/j.ccell.2019.05.005
Li, D., et al.: Estrogen-related hormones induce apoptosis by stabilizing schlafen-12 protein turnover. Mol. Cell 75(6), 1103–1116.e9 (2019). https://doi.org/10.1016/j.molcel.2019.06.040
Moroney, M.R., et al.: Inhibiting Wnt/beta-catenin in CTNNB1-mutated endometrial cancer. Mol. Carcinog. 60(8), 511–523 (2021). https://doi.org/10.1002/mc.23308
Hu, S., et al.: Molecular chaperones and Parkinson’s disease. Neurobiol. Dis. 160, 105527 (2021). https://doi.org/10.1016/j.nbd.2021.105527
Robinson, E.J., Aguiar, S., Smidt, M.P., van der Heide, L.P.: MCL1 as a therapeutic target in Parkinson’s disease? Trends Mol. Med. 25(12), 1056–1065 (2019). https://doi.org/10.1016/j.molmed.2019.08.009
Gelderblom, M., et al.: Temporal and spatial dynamics of cerebral immune cell accumulation in stroke. Stroke 40(5), 1849–57 (2009). https://doi.org/10.1161/STROKEAHA.108.534503
Nowinski, S.M., et al.: Mitochondrial fatty acid synthesis coordinates oxidative metabolism in mammalian mitochondria. Elife 9, e58041 (2020). https://doi.org/10.7554/eLife.58041
Valentijn, K., Valentijn, J.A., Jamieson, J.D.: Role of actin in regulated exocytosis and compensatory membrane retrieval: insights from an old acquaintance. Biochem. Biophys. Res. Commun. 266(3), 652–661 (1999). https://doi.org/10.1006/bbrc.1999.1883
Chen, R., et al.: Identification of JWA as a novel functional gene responsive to environmental oxidative stress induced by benzo[a]pyrene and hydrogen peroxide. Free Radic. Biol. Med. 42(11), 1704–1714 (2007). https://doi.org/10.1016/j.freeradbiomed.2007.02.018
Zhao, X., et al.: JWA antagonizes paraquat-induced neurotoxicity via activation of Nrf2. Toxicol. Lett. 277, 32–40 (2017). https://doi.org/10.1016/j.toxlet.2017.04.011
Li, X., Kumar, A., Carmeliet, P.: Metabolic pathways fueling the endothelial cell drive. Annu. Rev. Physiol. 81, 483–503 (2019). https://doi.org/10.1146/annurev-physiol-020518-114731
Gregorius, J., et al.: Small extracellular vesicles obtained from hypoxic mesenchymal stromal cells have unique characteristics that promote cerebral angiogenesis, brain remodeling and neurological recovery after focal cerebral ischemia in mice. Basic Res. Cardiol. 116(1), 40 (2021). https://doi.org/10.1007/s00395-021-00881-9
Acknowledgements
Thanks to all participants who have contributed to the study.
Decleration
Data Availability. The datasets selected in our research can be found and downloaded for free online, while the databases we searched and accession number(s) are all provided in article or supplement materials.
Funding.
This research received no external funding.
Author Contributions.
QYY was responsible for study conception; WWZ contributed to the methodology; YZ provided software; QYY wrote the original draft; YDZ contributed to the data analysis and supervised the execution of codes; YZ contributed to reviewing. All authors reviewed and approved the final version of the work.
Competing Interests.
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Yu, Q., Zhang, Y., Zhang, Y., Zhang, W. (2024). Diagnostic Genes Identification and Molecular Classification Patterns Based on Oxidative Stress-Related Genes in Ischemic Stroke. In: Su, R., Zhang, YD., Frangi, A.F. (eds) Proceedings of 2023 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2023). MICAD 2023. Lecture Notes in Electrical Engineering, vol 1166. Springer, Singapore. https://doi.org/10.1007/978-981-97-1335-6_17
Download citation
DOI: https://doi.org/10.1007/978-981-97-1335-6_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-1334-9
Online ISBN: 978-981-97-1335-6
eBook Packages: Computer ScienceComputer Science (R0)