Abstract
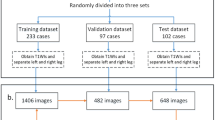
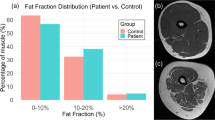
The diagnosis of specific types of muscular dystrophies (MD) is mainly done through genetic testing. As this does not always provide an unambiguous result, muscle MRI images are often examined to confirm or verify the diagnosis as each MD type affects the muscles in a specific pattern. Different deep learning approaches (ResNet50 model pretrained on RadImageNet, auto-encoder model trained from scratch, segmentation U-Net model trained for muscle segmentation) were investigated to obtain image features from Dixon MRI of the proximal leg that were used for discriminating between cases with Becker Muscular Dystrophy (n = 18), Limb-Girdle Muscular Dystrophy R12 (n = 13) or no MD (n = 16). The results are compared with classification by a conventional random forest (RF) classifier using the fat fraction percentage per muscle as features. The RF classifier and the segmentation U-Net deep learning approach performed best with an average AUC of 0.957 and 0.934 respectively. Local interpretable model-agnostic explanations (LIME) were used to explain the decisions of the RF model. Different fat replacement patterns for BMD and LGMDR12 observed in the glutei, adductors and vasti as described in literature were in part confirmed by the explanations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dıaz-Manera, J., Llauger, J., Gallardo, E., Illa, I.: Muscle MRI in muscular dystrophies. Acta Myologica XXXIV, 95–108 (2015)
Sarkozy, A., et al.: Muscle MRI findings in limb girdle muscular dystrophy type 2L. Neuromuscul. Disord. 22, S122−S129 (2012)
Tasca, G., et al.: Muscle MRI in Becker muscular dystrophy. Neuromuscul. Disord. 22 S100−S106 (2012)
Verdu-Dıaz, J., et al.: Accuracy of a machine learning muscle MRI-based tool for the diagnosis of muscular dystrophies
Mercuri, E., et al.: Clinical and imaging findings in six cases of congenital muscular dystrophy with rigid spine syndrome linked to chromosome 1p (RSMD1). Neuromuscul. Disord. 12, 631–638 (2002)
Ribeiro, M.T., Singh, S., Guestrin, C.: Why should I trust you? Explaining the predictions of any classifier. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1135–1144. Association for Computing Machinery (8 2016)
De Wel, B., et al.: Prospective natural history study in 24 adult patients with LGMDR12 over 2 years of follow-up: quantitative MRI and clinical outcome measures. Neurology 99(6), e638–e649 (2022)
Mei, X., et al.: RadImageNet: An open radiologic deep learning research dataset for effective transfer learning. Radiology: Artif. Intell. 4 (9 2022)
Kingma, D.P., Ba, J.: Adam: aA method for stochastic optimization (12 2014). http://arxiv.org/abs/1412.6980
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. 9351, 234–241 (2015)
Mercuri, E., et al.: Clinical and imaging findings in six cases of congenital muscular dystrophy with rigid spine syndrome linked to chromosome 1p (RSMD1). Neuromuscul. Disord. 12, 631–638 (2002)
Huysmans, L., De Wel, B., Claeys, K.G., Maes, F.: Automated MRI quantification of volumetric per-muscle fat fractions in the proximal leg of patients with muscular dystrophies. Front. Neurol. 14 1200727 (2023)
Acknowledgments
We thank the patients and healthy volunteers for their participation in the study. KGC is Chairholder of the Emil von Behring Chair for Neuromuscular and Neurodegenerative Disorders by CSL Behring. KGC is member of the European Reference Network for Rare Neuromuscular Diseases (ERN EURO-NMD) and of the European Reference Network for Rare Neurological Diseases (ERN-RND). The authors report no disclosures relevant to the manuscript.
Funding
This work is supported in part by the Internal Funds KU Leuven under Grant C24/18/047 and by the Flemish Government under the “Onderzoeksprogramma Artificiële Intelligentie (AI) Vlaanderen” programme. BDW is supported by the Fund for Scientific Research Flanders (FWO, PhD fellowship fundamental research grant number 1159121N).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Huysmans, L., De Wel, B., Iterbeke, L., Claeys, K., Maes, F. (2024). Deep Learning Approaches for Automated Classification of Muscular Dystrophies from MRI. In: Su, R., Zhang, YD., Frangi, A.F. (eds) Proceedings of 2023 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2023). MICAD 2023. Lecture Notes in Electrical Engineering, vol 1166. Springer, Singapore. https://doi.org/10.1007/978-981-97-1335-6_24
Download citation
DOI: https://doi.org/10.1007/978-981-97-1335-6_24
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-1334-9
Online ISBN: 978-981-97-1335-6
eBook Packages: Computer ScienceComputer Science (R0)