Abstract
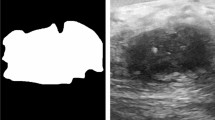
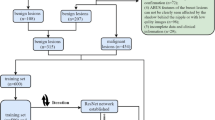
Breast cancer is still the predominant type of cancer that affects women worldwide. Artificial intelligence (AI) developers are making great efforts in developing automated computer-aided detection and diagnosis (CAD) systems for breast cancer prognosis and classification. There are currently no official guidelines recommending the use of AI with ultrasound in clinical practice, and further research is needed to investigate more advanced approaches and demonstrate their utility. In this paper, we use one of the state-of-the-arts segmentation model based on deep learning, as well as compare different recent pre-trained models for the purpose of segmenting and distinguishing between benign and malignant masses in breast ultrasound images. The results showed that among the compared models, the most accurate model was the EfficientNetB7 model which achieved 88% on the segmented input images, that were segmented using the ResUNet model with 0.8932 Dice score and 0.8572 Iou score.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
World Health Organization: Breast cancer web site. https://www.who.int/news-room/fact-sheets/detail/breast-cancer (Accessed 20 April 2023)
Sennoga, C.A.: Ultrasound imaging. Bioengineering Innovative Solutions for Cancer. Elsevier Ltd. (2019). https://doi.org/10.1016/B978-0-12-813886-1.00007-3
Giger, M.L.: Computer-aided diagnosis in diagnostic mammography and multimodality breast imaging. RSNA Categ. Course Diagnostic Radiol. Phys. Adv. Breast Imaging—Physics, Technol. Clin. Appl., 205–217 (2004)
Xiao, T., et al.: Comparison of transferred deep neural networks in ultrasonic breast masses discrimination. Biomed. Res. Int. 2018, 4605191 (2018)
Sun, Q., et al.: Deep Learning vs. Radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don’t forget the peritumoral region. Front. Oncol. 10, 53 (2020)
Li, C., et al.: A convolutional neural network based on ultrasound images of primary breast masses: prediction of lymph-node metastasis in collaboration with classification of benign and malignant tumors. Front. Physiol. 13 (2022)
Liao, W.-X., et al.: Automatic identification of breast ultrasound image based on supervised block-based region segmentation algorithm and features combination migration deep learning model. IEEE J. Biomed. Heal. Inform. 24, 984–993 (2020)
Vakanski, A., Xian, M., Freer, P.E.: Attention-enriched deep learning model for breast tumor segmentation in ultrasound images. Ultrasound Med. Biol. 46, 2819–2833 (2020)
Shorten, C., Khoshgoftaar, T.M.: A survey on image data augmentation for deep learning. J. Big Data 6, 60 (2019)
Krizhevsky, A., Sutskever, I., E. Hinton, G.: ImageNet classification with deep convolutional neural networks. Neural Inform. Proc. Syst. 25 (2012)
Simonyan, K., Zisserman, A.: Very Deep Convolutional Networks for Large-Scale Image Recognition (2014). https://doi.org/10.48550/ARXIV.1409.1556
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016). doi:https://doi.org/10.1109/CVPR.2016.90
Huang, G., Liu, Z., Maaten, L. Van Der & Weinberger, K. Q. Densely connected convolutional networks. in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2261–2269 (2017). https://doi.org/10.1109/CVPR.2017.243
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Hijab, A., Rushdi, M.A., Gomaa, M.M., Eldeib, A.: breast cancer classification in ultrasound images using transfer learning. In: International Conference on Advanced Bioinformatics and Biomedical Engineering, ICABME 2019, pp. 1–4 (2019)
Baccouche, A., Garcia-Zapirain, B., Castillo Olea, C., Elmaghraby, A.S.: Connected-UNets: a deep learning architecture for breast mass segmentation. npj Breast Cancer 7, 1–12 (2021)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: Convolutional Networks for Biomedical Image Segmentation (2015). https://doi.org/10.48550/ARXIV.1505.04597
Punn, N.S., Agarwal, S.: RCA-IUnet: a residual cross-spatial attention-guided inception U-Net model for tumor segmentation in breast ultrasound imaging. Mach. Vis. Appl. 33, 27 (2022)
Al-Dhabyani, W., Gomaa, M., Khaled, H., Fahmy, A.: Dataset of breast ultrasound images. Data in Brief. 28, 104863 2020). https://doi.org/10.1016/j.dib.2019.104863. https://scholar.cu.edu.eg/?q=afahmy/pages/dataset (Accessed 15 Dec 2021)
Gokhale, S.: Ultrasound characterization of breast masses. Indian J. Radiol. Imaging 19, 242–247 (2009)
Google Colaboratory. https://colab.research.google.com/ (Accessed 20 April 2023)
Acknowledgment
All authors gratefully acknowledged the Egyptian Academy of Scientific Research and Technology (ASRT) for providing financial support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Ekhlas, S., Abd-Elsalam, N.M., AlSaidy, Z.A., Kandil, A.H., Al-bialy, A., Youssef, A.B.M. (2024). Comparing Different Deep-Learning Models for Classifying Masses in Ultrasound Images. In: Su, R., Zhang, YD., Frangi, A.F. (eds) Proceedings of 2023 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2023). MICAD 2023. Lecture Notes in Electrical Engineering, vol 1166. Springer, Singapore. https://doi.org/10.1007/978-981-97-1335-6_28
Download citation
DOI: https://doi.org/10.1007/978-981-97-1335-6_28
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-1334-9
Online ISBN: 978-981-97-1335-6
eBook Packages: Computer ScienceComputer Science (R0)