Abstract
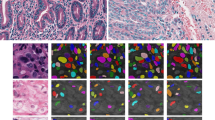
Chromogenic RNAscope dye and haematoxylin staining of cancer tissue facilitates diagnosis of the cancer type and subsequent treatment, and fits well into existing pathology workflows. However, manual quantification of the RNAscope transcripts (dots), which signify gene expression, is prohibitively time consuming. In addition, there is a lack of verified supporting methods for quantification and analysis. This paper investigates the usefulness of gray level texture features for automatically segmenting and classifying the positions of RNAscope transcripts from breast cancer tissue. Feature analysis showed that a small set of gray level features, including Gray Level Dependence Matrix and Neighbouring Gray Tone Difference Matrix features, were well suited for the task. The automated method performed similarly to expert annotators at identifying the positions of RNAscope transcripts, with an \(F_1\)-score of 0.571 compared to the expert inter-rater \(F_1\)-score of 0.596. These results demonstrate the potential of gray level texture features for automated quantification of RNAscope in the pathology workflow.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Amadasun, M., King, R.: Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 19(5), 1264–1274 (1989). https://doi.org/10.1109/21.44046
Arnold, M., et al.: Current and future burden of breast cancer: global statistics for 2020 and 2040. The Breast 66, 15–23 (2022). https://doi.org/10.1016/j.breast.2022.08.010
Chowdhary, C.L., Acharjya, D.: Segmentation and feature extraction in medical imaging: a systematic review. Procedia Comp. Sci. 167, 26–36 (2020). International Conference on Computational Intelligence and Data Science. https://doi.org/10.1016/j.procs.2020.03.179
Galloway, M.M.: Texture analysis using gray level run lengths. Comput. Graph. Image Process. 4(2), 172–179 (1975). https://doi.org/10.1016/S0146-664X(75)80008-6
Ghaznavi, F., Evans, A., Madabhushi, A., Feldman, M.: Digital imaging in pathology: whole-slide imaging and beyond. Annu. Rev. Pathol. 8(1), 331–359 (2013). https://doi.org/10.1146/annurev-pathol-011811-120902
Graschew, G., Roelofs, T.A., Rakowsky, S., Schlag, P.M.: E-health and telemedicine. Int. J. Comput. Assist. Radiol. Surg. 1(1), 119–135 (2006). https://doi.org/10.1007/s11548-006-0012-1
van Griethuysen, J.J., et al.: Computational radiomics system to decode the radiographic phenotype. Can. Res. 77(21), e104–e107 (2017). https://doi.org/10.1158/0008-5472.CAN-17-0339
Haralick, R.M., Shanmugam, K., Dinstein, I.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC- 3(6), 610–621 (1973). https://doi.org/10.1109/tsmc.1973.4309314
Morley-Bunker, A.E., et al.: RNAscope compatibility with image analysis platforms for the quantification of tissue-based colorectal cancer biomarkers in archival formalin-fixed paraffin-embedded tissue. Acta Histochemica 123(6), 151, 765 (2021). https://doi.org/10.1016/j.acthis.2021.151765
Rozario, S.Y., Sarkar, M., Farlie, M.K., Lazarus, M.D.: Responding to the healthcare workforce shortage: a scoping review exploring anatomical pathologists’ professional identities over time. Anat. Sci. Educ. 17, 351–365 (2023). https://doi.org/10.1002/ase.2260
Ruifrok, A.C., Johnston, D.A.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23(4), 291–299 (2001)
Sun, C., Wee, W.G.: Neighboring gray level dependence matrix for texture classification. Comput. Vis. Graph. Image Process. 23(3), 341–352 (1983). https://doi.org/10.1016/0734-189X(83)90032-4
Suzuki, S., Be, K.: Topological structural analysis of digitized binary images by border following. Comput. Vis. Graph Image Process. 30(1), 32–46 (1985). https://doi.org/10.1016/0734-189X(85)90016-7
Thibault, G., et al.: Texture indexes and gray level size zone matrix application to cell nuclei classification. In: International Conference on Pattern Recognition and Information Processing, PRIP 2009, pp. 140–145 (2009)
Wang, F., et al.: RNAScope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. JMD 14(1), 22–29 (2012). https://doi.org/10.1016/j.jmoldx.2011.08.002
Acknowledgment
This study used data supplied by Associate Professor Logan Walker, Dr. Arthur Morley-Bunker, and Dr. George Wiggins from the University of Otago, Christchurch.
The RNAscope transcripts on a total of 144 image patches were annotated by Dr. Arthur Morley-Bunker (a trained pathology scientist) and Dr. Gavin Harris (an anatomical pathologist) for use in this study.
We wish to thank Heather Thorne, Eveline Niedermayr, Sharon Guo, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia.
Author information
Authors and Affiliations
Consortia
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Davidson, A. et al. (2024). Grey Level Texture Features for Segmentation of Chromogenic Dye RNAscope from Breast Cancer Tissue. In: Su, R., Zhang, YD., Frangi, A.F. (eds) Proceedings of 2023 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2023). MICAD 2023. Lecture Notes in Electrical Engineering, vol 1166. Springer, Singapore. https://doi.org/10.1007/978-981-97-1335-6_7
Download citation
DOI: https://doi.org/10.1007/978-981-97-1335-6_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-1334-9
Online ISBN: 978-981-97-1335-6
eBook Packages: Computer ScienceComputer Science (R0)