Abstract
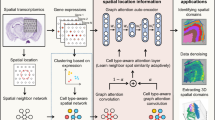
Exploring spatial domains to investigate tissue structures is a chance provided by spatial transcriptome technology, while also a significant challenge in spatial transcriptomics research. Current approaches only focus on spatial gene expression and cannot simultaneously incorporate spatial location information. Graph deep learning models can simultaneously encode node features and positional information. However, during decoding, most of the models still only focus on reconstructing feature information, ignoring positional information. Here, we propose a new method, DeepDomain, which aims to improve the latent representation of nodes by jointly reconstructing gene expression profiles and spatial neighborhood networks using a deep graph attention network with two distinct decoders. Utilizing enhanced spatial latent representations to identify spatial domains in three datasets, DeepDomain achieved higher accuracy in evaluation metrics and a better description of organizational structure when compared to existing methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Rao, A., Barkley, D., Franca, G.S., Yanai, I.: Exploring tissue architecture using spatial transcriptomics. Nature 596(7871), 211–220 (2021). https://doi.org/10.1038/s41586-021-03634-9
Wang, X., et al.: Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science (New York, NY) 361(6400) (2018). https://doi.org/10.1126/science.aat5691
Baron M., et al.: The stress-like cancer cell state is a consistent component of tumorigenesis. Cell Syst. 11(5), 536–46.e7 (2020). https://doi.org/10.1016/j.cels.2020.08.018
Moncada, R., et al.: Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nature Biotechnol. 38(3), 333–42 (2020). https://doi.org/10.1038/s41587-019-0392-8
Chen, J., Xu, H., Tao, W., Chen, Z., Zhao, Y., Han, J.J.: Transformer for one stop interpretable cell type annotation. Nat. Commun. 14(1), 223 (2023). https://doi.org/10.1038/s41467-023-35923-4
Li, X., et al.: Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat. Commun. 11(1), 2338 (2020). https://doi.org/10.1038/s41467-020-15851-3
Hu, J., et al.: SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18(11), 1342–1351 (2021). https://doi.org/10.1038/s41592-021-01255-8
Dong, K., Zhang, S.: Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. Nat. Commun. 13(1), 1739 (2022). https://doi.org/10.1038/s41467-022-29439-6
Yongshuo, Z., Tingyang, Y., Xuesong, W., Yixuan, W., Zhihang, H, Yu L.: conST: an interpretable multi-modal contrastive learning framework for spatial transcriptomics. bioRxiv. 2022.01.14.476408 (2022). https://doi.org/10.1101/2022.01.14.476408
Ren, H., Walker, B.L., Cang, Z., Nie, Q.: Identifying multicellular spatiotemporal organization of cells with SpaceFlow. Nat. Commun. 13(1), 4076 (2022). https://doi.org/10.1038/s41467-022-31739-w
Zhao, E., Stone, M.R., Ren, X., Guenthoer, J., Smythe, K.S., Pulliam, T., et al.: Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39(11), 1375–1384 (2021). https://doi.org/10.1038/s41587-021-00935-2
Velickovic, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., Bengio, Y.J.S.: Graph Attention Netw. 1050(20), 10–48550 (2017)
Scrucca, L., Fop, M., Murphy, T.B., Raftery, A.E.: Mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J. 8(1), 289–317 (2016)
Maynard, K.R., et al.: Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24(3), 425–436 (2021). https://doi.org/10.1038/s41593-020-00787-0
Wolf, F.A., Angerer, P., Theis, F.J.: SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19(1), 15 (2018). https://doi.org/10.1186/s13059-017-1382-0
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). https://doi.org/10.48550/arXiv.1201.0490
Satija, R., Farrell, J.A., Gennert, D., Schier, A.F., Regev, A.: Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33(5), 495–502 (2015). https://doi.org/10.1038/nbt.3192
Acknowledgments
The work was supported by the National Natural Science Foundation of China (No. 62131004), the National Key R&D Program of China (2022ZD0117700).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Liu, Y., Zou, Q. (2024). Spatial Domain Identifying: Graph Attention Network with Two Different Decoders. In: Huang, DS., Zhang, Q., Guo, J. (eds) Advanced Intelligent Computing in Bioinformatics. ICIC 2024. Lecture Notes in Computer Science(), vol 14881. Springer, Singapore. https://doi.org/10.1007/978-981-97-5689-6_27
Download citation
DOI: https://doi.org/10.1007/978-981-97-5689-6_27
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-5688-9
Online ISBN: 978-981-97-5689-6
eBook Packages: Computer ScienceComputer Science (R0)