Abstract
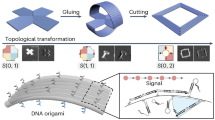
Due to its high degree of customization, DNA origami provides a versatile platform with which to engineer nanoscale structures and devices. Reconfigurable nanodevices driven by DNA strand displacement accomplish the task of transition between different conformations, endowing DNA origami with application values. Herein, we propose a strategy to regulate the conformation of DNA origami using the polymerase-triggered DNA strand displacement (PTSD) reaction. We design a book-shaped DNA origami structure consisting of four pages connected into a cuboid shape. The PTSD reactions initiated by different primer strands selectively remove the connecting strand, transforming the nano book into a two-page or a four-page conformation. We utilize three primer strands to remove thirty-five connecting strands and construct three conformations of the identical DNA origami, illustrating that the PTSD reaction is an effective tool for the reconfiguration of DNA origami. The statistical results of TEM images prove the effectiveness of the proposed method. Our work on the development of PTSD-driven reconfigurable nanostructure will offer a new way to create intelligent materials for advanced nanotechnology applications.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Rothemund, P.W.K.: Folding DNA to create nanoscale shapes and patterns. Nature 440(7082), 297–302 (2006)
Liu, W., Zhong, H., Wang, R., Seeman, N.C.: Crystalline two-dimensional DNA-origami arrays. Angew. Chem. Int. Ed. 50(1), 264–267 (2011)
Woo, S., Rothemund, P.W.K.: Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3(8), 620–627 (2011)
Han, D., Pal, S., Nangreave, J., Deng, Z., Liu, Y., Yan, H.: DNA origami with complex curvatures in three-dimensional space. Science 332(6027), 342–346 (2011)
Douglas, S.M., Dietz, H., Liedl, T., Högberg, B., Graf, F., Shih, W.M.: Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459(7245), 414–418 (2009)
Ke, Y., et al.: Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 131(43), 15903–15908 (2009)
Kopperger, E., List, J., Madhira, S., Rothfischer, F., Lamb, D.C., Simmel, F.C.: A self-assembled nanoscale robotic arm controlled by electric fields. Science 359(6373), 296–301 (2018)
Li, S., et al.: A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36(3), 258–264 (2018)
Ke, Y., Meyer, T., Shih, W.M., Bellot, G.: Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat. Commun. 7(1), 10935 (2016)
Kuzyk, A., Yang, Y., Duan, X., Stoll, S., Govorov, A.O., Sugiyama, H., Endo, M., Liu, N.: A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun. 7(1), 10591 (2016)
Chen, K., Zhu, J., Bošković, F., Keyser, U.F.: Nanopore-based DNA hard drives for rewritable and secure data storage. Nano Lett. 20(5), 3754–3760 (2020)
Song, J., Li, Z., Wang, P., Meyer, T., Mao, C., Ke, Y.: Reconfiguration of DNA molecular arrays driven by information relay. Science 357(6349), eaan3377 (2017)
Gu, H., Chao, J., Xiao, S.-J., Seeman, N.C.: A proximity-based programmable DNA nanoscale assembly line. Nature 465(7295), 202–205 (2010)
Kuzuya, A., Sakai, Y., Yamazaki, T., Xu, Y., Komiyama, M.: Nanomechanical DNA origami “single-molecule beacons’’ directly imaged by atomic force microscopy. Nature Commun. 2(1), 449 (2011)
Gerling, T., Wagenbauer, K.F., Neuner, A.M., Dietz, H.: Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347(6229), 1446–1452 (2015)
Ryssy, J., et al.: Light-responsive dynamic DNA-origami-based plasmonic assemblies. Angew. Chem. Int. Ed. 60(11), 5859–5863 (2021)
Chen, Z., Chen, K., Xie, C., Liao, K., Xu, F., Pan, L.: Cyclic transitions of DNA origami dimers driven by thermal cycling. Nanotechnology 34(6), 065601 (2023)
Ijäs, H., Hakaste, I., Shen, B., Kostiainen, M.A., Linko, V.: Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 13(5), 5959–5967 (2019)
Douglas, S.M., Bachelet, I., Church, G.M.: A logic-gated nanorobot for targeted transport of molecular payloads. Science 335(6070), 831–834 (2012)
Han, D., Pal, S., Liu, Y., Yan, H.: Folding and cutting DNA into reconfigurable topological nanostructures. Nat. Nanotechnol. 5(10), 712–717 (2010)
Chen, K., Xu, F., Hu, Y., Yan, H., Pan, L.: DNA kirigami driven by polymerase-triggered strand displacement. Small 18(24), 2201478 (2022)
Liao, K., Chen, K., Xie, C., Chen, Z., Pan, L.: Disassembly of DNA origami dimers controlled by programmable polymerase primers. Chem. Commun. 58(92), 12879–12882 (2022)
Acknowledgment
This work was supported by National Natural Science Foundation of China (62172171), Zhejiang Lab (2021RD0AB03), and Fundamental Research Funds for the Central Universities (HUST: 2019kfyXMBZ056).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Chen, K., Chen, Z., Xie, C., Pan, L. (2023). Reconfigurable Nanobook Structure Driven by Polymerase-Triggered DNA Strand Displacement. In: Pan, L., Zhao, D., Li, L., Lin, J. (eds) Bio-Inspired Computing: Theories and Applications. BIC-TA 2022. Communications in Computer and Information Science, vol 1801. Springer, Singapore. https://doi.org/10.1007/978-981-99-1549-1_54
Download citation
DOI: https://doi.org/10.1007/978-981-99-1549-1_54
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-1548-4
Online ISBN: 978-981-99-1549-1
eBook Packages: Computer ScienceComputer Science (R0)