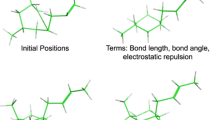
Summary
This paper describes the further development of the functionality of our in-house de novo design program, PRO_LIGAND. In particular, attention is focussed on the implementation and validation of the ‘directed tweak’ method for the construction of conformationally flexible molecules, such as peptides, from molecular fragments. This flexible fitting method is compared to the original method based on libraries of prestored conformations for each fragment. It is shown that the directed tweak method produces results of comparable quality, with significant time savings. By removing the need to generate a set of representative conformers for any new library fragment, the flexible fitting method increases the speed and simplicity with which new fragments can be included in a fragment library and also reduces the disk space required for library storage. A further improvement to the molecular construction process within PRO_LIGAND is the inclusion of a constrained minimisation procedure which relaxes fragments onto the design model and can be used to reject highly strained structures during the structure generation phase. This relaxation is shown to be very useful in simple test cases, but restricts diversity for more realistic examples. The advantages and disadvantages of these additions to the PRO_LIGAND methodology are illustrated by three examples: similar design to an alpha helix region of dihydrofolate reductase, complementary design to the active site of HIV-1 protease and similar design to an epitope region of lysozyme.
Similar content being viewed by others
References
Verlinde, C.L.M.J. and Hol, W.G.J., Structure, 2 (1994) 577.
Lewis, R.A. and Leach, A.R., J. Comput.-Aided Mol. Design, 8 (1994) 467.
Nishibata, Y. and Itai, A., Tetrahedron, 47 (1991) 8985.
Nishibata, Y. and Itai, A., J. Med. Chem., 36 (1993) 2921.
Rotstein, S.H. and Murcko, M.A., J. Comput.-Aided Mol. Design, 7 (1993) 23.
Pearlman, D.A. and Murcko, M.A., J. Comput. Chem., 14 (1993) 1184.
Gehlhaar, D.K., Moerder, K.E., Zichi, D., Sherman, C.J., Ogden, R.C. and Freer, S.T., J. Med. Chem., 38 (1995) 466.
Moon, J.B. and Howe, W.J., Protein Struct. Funct. Genet., 11 (1991) 314.
Moon, J.B. and Howe, W.J., In Wermuth, C.G. (Ed.) Trends in QSAR and Molecular Modelling 92 (Proceedings of the 9th European Symposium on Structure-Activity Relationships: QSAR and Molecular Modelling), ESCOM, Leiden, 1993, pp. 11–19.
Miranker, A. and Karplus, M., Protein Struct. Funct. Genet., 11 (1991) 29.
Caflisch, A., Miranker, A. and Karplus, M., J. Med. Chem., 36 (1993) 2142.
Rotstein, S.H. and Murcko, M.A., J. Med. Chem., 36 (1993) 1700.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 6 (1992) 61.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 6 (1992) 593.
Böhm, H.-J., In Kubinyi, H. (Ed.) 3D QSAR in Drug Design: Theory, Methods and Applications, ESCOM, Leiden, 1993, pp. 386–405.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 8 (1994) 243.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 8 (1994) 623.
Lewis, R.A., Roe, D.C., Huang, C., Ferrin, T.E., Langridge, R. and Kuntz, I.D., J. Mol. Graphics, 10 (1992) 66.
Roe, D.C. and Kuntz, I.D., J. Comput.-Aided Mol. Design, 9 (1995) 269.
Tschinke, V. and Cohen, N.C., J. Med. Chem., 36 (1993) 3863.
Ho, C.W.M. and Marshall, G.R., J. Comput.-Aided Mol. Design, 7 (1993) 623.
Leach, A.R. and Kilvington, S.R., J. Comput.-Aided Mol. Design, 8 (1994) 283.
Eisen, M.B., Wiley, D.C., Karplus, M. and Hubbard, R.E., Protein Struct. Funct. Genet., 19 (1994) 199.
Bohacek, R.S. and McMartin, C., J. Am. Chem. Soc., 116 (1994) 5560.
Clark, D.E., Frenkel, D., Levy, S.A., Li, J., Murray, C.W., Robson, B., Waszkowycz, B. and Westhead, D.R., J. Comput.-Aided Mol. Design, 9 (1995) 13.
Waszkowycz, B., Clark, D.E., Frenkel, D., Li, J., Murray, C.W., Robson, B. and Westhead, D.R., J. Med. Chem., 37 (1994) 3994.
Westhead, D.R., Clark, D.E., Frenkel, D., Li, J., Murray, C.W., Robson, B. and Waszkowycz, B., J. Comput.-Aided Mol. Design, 9 (1995) 139.
Frenkel, D., Clark, D.E., Li, J., Murray, C.W., Robson, B., Waszkowycz, B. and Westhead, D.R., J. Comput.-Aided Mol. Design, 9 (1995) 213.
Gillet, V.J., Johnson, A.P., Mata, P., Sike, S. and Williams, P., J. Comput.-Aided Mol. Design, 7 (1993) 127.
Gillet, V.J., Newell, W., Mata, P., Myatt, G., Sike, S., Zsoldos, Z. and Johnson, A.P., J. Chem. Inf. Comput. Sci., 34 (1994) 207.
Mata, P., Gillet, V.J., Johnson, A.P., Lampreia, J., Myatt, G.J., Sike, S. and Stebbings, A.L., J. Chem. Inf. Comput. Sci., 35 (1995) 479.
Leach, A.R. and Lewis, R.A., J. Comput. Chem., 15 (1994) 233.
Cohen, A.A. and Shatzmiller, S.E., J. Mol. Graphics, 11 (1993) 166.
Cohen, A.A. and Shatzmiller, S.E., J. Comput. Chem., 15 (1994) 1393.
Glen, R.C. and Payne, A.W.R., J. Comput.-Aided Mol. Design, 9 (1995) 181.
Clark, D.E. and Murray, C.W., J. Chem. Inf. Comput. Sci., in press.
Smellie, A., Kahn, S. and Teig, S.L., J. Chem. Inf. Comput. Sci., 35 (1995) 295.
Smellie, A., Kahn, S. and Teig, S.L., J. Chem. Inf. Comput. Sci., 35 (1995) 295.
Pearlman, R.S., In Kubinyi, H. (Ed.) 3D QSAR in Drug Design: Theory, Methods and Applications, ESCOM, Leiden, 1993, pp. 41–79.
Clark, D.E., Jones, G., Willett, P., Kenny, P.W. and Glen, R.C., J. Chem. Inf. Comput. Sci., 34 (1994) 197.
Moock, T.E., Henry, D.R., Ozkabak, A.G. and Alamgir, M., J. Chem. Inf. Comput. Sci., 34 (1994) 184.
Hurst, T., J. Chem. Inf. Comput. Sci., 34 (1994) 190.
Shenkin, P.S., Yarmush, D.L., Fine, R.M., Wang, H. and Levinthal, C., Biopolymers, 26 (1987) 2053.
Fine, R.M., Wang, H., Shenkin, P.S., Yarmush, D.L. and Levinthal, C., Protein Struct. Funct. Genet., 1 (1986) 342.
Weiner, S.J., Kollman, P.A., Case, D.A., Singh, U.C., Ghio, C., Alagona, G., Profeta, Jr., S. and Weiner, P., J. Am. Chem. Soc., 106 (1984) 765.
Siani, M.A., Weininger, D. and Blaney, J.M., J. Chem. Inf. Comput. Sci., 34 (1994) 588.
Gallop, M.A., Barrett, R.W., Dower, W.J., Fodor, S.P.A. and Gordon, E.M., J. Med. Chem., 37 (1994) 1233.
Gordon, E.M., Barrett, R.W., Dower, W.J., Fodor, S.P.A. and Gallop, M.A., J. Med. Chem., 37 (1994) 1385.
Klebe, G., J. Mol. Biol., 237 (1994) 212.
Allen, F.H., Bellard, S., Brice, M.D., Cartwright, B.A., Doubleday, A., Higgs, H., Hummelink, T., Hummelink-Peters, B.G., Kennard, O., Motherwell, W.D.W., Rodgers, J.R. and Watson, D.G., Acta Crystallogr., B35 (1979) 2331.
Ullmann, J.R., J. Assoc. Comput. Machinery, 23 (1976) 31.
Brint, A.T. and Willett, P., J. Mol. Graphics, 5 (1987) 49.
Clark, D.E., Willett, P. and Kenny, P.W., J. Mol. Graphics, 10 (1992) 194.
Murrall, N.W. and Davies, E.K., J. Chem. Inf. Comput. Sci., 30 (1990) 312.
Wenger, J.C. and Smith, D.H., J. Chem. Inf. Comput. Sci., 22 (1982) 29.
Dress, A.W.M. and Havel, T.F., Discrete Applied Math., 19 (1988) 129.
Jordan, S., Leach, A.R. and Bradshaw, J., J. Chem. Inf. Comput. Sci., 35 (1995) 640.
Insight II, v. 2.3.0, Biosym Technologies, Inc., San Diego, CA.
Bolin, J.T., Filman, D.J., Matthews, D.A., Hamlin, R.C. and Kraut, J., J. Biol. Chem., 257 (1982) 13650.
Appelt, K., Perspect. Drug Discov. Design, 1 (1993) 23.
Fitzgerald, P.M.D., Curr. Opin. Struct. Biol., 3 (1993) 868.
Redshaw, S., Exp. Opin. Invest. Drugs, 3 (1994) 273.
Fitzgerald, P.M.D., McKeever, B.M., Van, Middlesworth, J.F., Springer, J.P., Heimbach, J.C., Leu, C.-T., Herber, W.K., Dixon, R.A.F. and Darke, P.L., J. Biol. Chem., 265 (1990) 14209.
Fischmann, T.O., Bentley, G.A., Bhat, T.N., Boulot, G., Mariuzza, R.A., Phillips, S.E.V., Tello, D. and Poljak, R.J., J. Biol. Chem., 266 (1991) 12915.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Murray, C.W., Clark, D.E. & Byrne, D.G. PRO_LIGAND: An approach to de novo molecular design. 6. Flexible fitting in the design of peptides. J Computer-Aided Mol Des 9, 381–395 (1995). https://doi.org/10.1007/BF00123996
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00123996