Summary
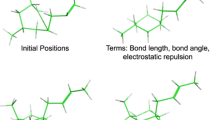
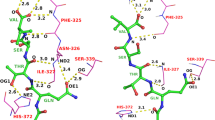
SPLICE is a program that processes partial query solutions retrieved from 3D, structural databases to generate novel, aggregate ligands. It is designed to interface with the database searching program FOUNDATION, which retrieves fragments containing any combination of a user-specified minimum number of matching query elements. SPLICE eliminates aspects of structures that are physically incapable of binding within the active site. Then, a systematic rule-based procedure is performed upon the remaining fragments to ensure receptor complementarity. All modifications are automated and remain transparent to the user. Ligands are then assembled by linking components into composite structures through overlapping bonds. As a control experiment, FOUNDATION and SPLICE were used to reconstruct a know HIV-1 protease inhibitor after it had been fragmented, reoriented, and added to a sham database of fifty different small molecules. To illustrate the capabilities of this program, a 3D search query containing the pharmacophoric elements of an aspartic proteinase-inhibitor crystal complex was searched using FOUNDATION against a subset of the Cambridge Structural Database. One hundred thirty-one compounds were retrieved, each containing any combination of at least four query elements. Compounds were automatically screened and edited for receptor complementarity. Numerous combinations of fragments were discovered that could be linked to form novel structures, containing a greater number of pharmacophoric elements than any single retrieved fragment.
Similar content being viewed by others
References
Martin, Y.C., J. Med. Chem., 35 (1992) 2145.
Martin, Y.C., Bures, M.G. and Willett, P., In Lipkowitz, K.B. and Boyd, D.B. (Eds.) Reviews in Computational Chemistry, VCH Publishers, Inc., New York, 1990, pp. 213–256.
Murrall, N.W. and Davies, E.K., J. Chem. Inf. Comput. Sci., 30 (1990) 312.
Fersht, A., Enzyme Structure and Mechanism, Freeman, New York, 1977.
Beddell, C.R., The Design of Drugs to Macromolecular Targets, Wiley, New York, 1992.
DesJarlais, R.L., Sheridan, R.P., Dixon, J.S., Kuntz, I.D. and Venkataraghavan, R.J., J. Med. Chem., 29 (1986) 2149.
Kuntz, I.D., Blaney, J.M., Oatley, S.J., Langridge, R. and Ferrin, T.E., J. Mol. Biol., 161 (1982) 269.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 6 (1992) 61.
Böhm, H.-J., J. Comput.-Aided Mol. Design, 6 (1992) 593.
Chau, P.L. and Dean, P.M., J. Comput.-Aided Mol. Design, 6 (1992) 385.
Chau, P.L. and Dean, P.M., J. Comput.-Aided Mol. Design, 6 (1992) 397.
Chau, P.L. and Dean, P.M., J. Comput.-Aided Mol. Design, 6 (1992) 407.
Allen, F.H., Kennard, O. and Taylor, R., Acc. Chem. Res., 16 (1983) 146.
Miranker, A. and Karplus, M., Protein Struct. Funct. Genet., 11 (1991) 29.
Ho, C.M.W. and Marshall, G.R., J. Comput.-Aided Mol. Design, 7 (1993) 3.
Lewis, R.A. and Dean, P.M., Proc. Roy. Soc. London Ser. B., 236 (1989) 125.
Lewis, R.A. and Dean, P.M., Proc. Roy. Soc. London Ser. B., 236 (1989) 141.
Lewis, R.A., Roe, D.C., Huang, C., Ferrin, T.E., Langridge, R. and Kuntz, I.D., J. Mol. Graphics, 10 (1992) 66.
Lewis, R.A. and Kuntz, I.D., In Karalainen, E.J. (Ed.) Scientific Computering and Automation, Elsevier, Amsterdam, 1991, pp. 117–132.
Cormen, T.H., Leiserson, C.E. and Rivest, R.L., Introduction to Algorithms, McGraw-Hill Book Co., St. Louis, 1991.
Tripos Associates, Inc., St. Louis, MO, U.S.A.
Rich, D.H., Green, J., Toth, M.V., Marshall, G.R. and Kent, S.B.H., J. Med. Chem., 33 (1990) 1285.
Suguna, K., Padlan, E.A., Smith, C.W., Carlson, W.D. and Davies, D.R., Proc. Natl. Acad. Sci. USA, 84 (1987) 7009.
Ho, C.M.W. and Marshall, G.R., J. Comput.-Aided Mol. Design, 4 (1990) 337.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ho, C.M.W., Marshall, G.R. SPLICE: A program to assemble partial query solutions from three-dimensional database searches into novel ligands. J Computer-Aided Mol Des 7, 623–647 (1993). https://doi.org/10.1007/BF00125322
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00125322