Abstract
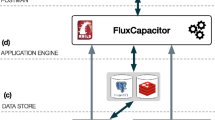
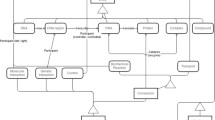
The requirements of a traditional integration of databases include hiding the heterogeneity among its member databases, preserving their autonomy, supporting controlled data sharing, and providing a user-friendly interface, with a performance comparable to that of a homogeneous distributed database system. In this paper, we describe a variation of the loosely coupled federated database approach that satisfies these requirements and is specifically oriented toward the special characteristics and needs of molecular biology databases. The approach differs from the traditional federated database approach in that it also integrates software tools together with the databases. In addition, the system gives the graphical user interface a more central role in the integration. Thus, the system has all the advantages provided by a visual user environment and is thus more user-friendly than traditional linguistic approaches. The paper also includes a description of XBio, a system which constructs such an integration. The design of XBio ensures robustness and can be customized to the needs of different classes of users.
Similar content being viewed by others
References
S.F. Altschul et al., “A basic alignment search tool,”J. Mol. Biol., vol. 215, p. 403, 1990.
H.S. Bilofsky et al., “The GenBank genetic sequence databank,”Nucleic Acid Res., vol. 14, pp. 1–17, 1985.
J. Devereux, P. Haeberli, and O. Smithies, “A comprehensive set of sequence analysis programs for the VAX,”Nucleic Acid Res. vol. 12, p. 387, 1984.
C.J. Gartmann and U. Grob, “A menu-shell for the GCG programs,”CABIOS, vol. 7, no. 4, 1991.
G.H. Gonnet and F.W. Tompa, “Mind your grammar: a new approach to modeling text,”Proc. 13th VLDB Conf., Brighton, England, 1987, pp. 339–346.
K. Hammer and D. Mcleod, “On database management system architecture,”Tech. Reports, MIT/LCS/TM-141, 1979.
N.N. Kamel, “A profile for molecular biology databases,”Comput. Appl. BOsci.
N.N. Kamel and M. Kamel, “Federated database management system: requirements, issues, and solutions,”Computer Communications, vol. 15, no. 6, pp. 270–278, 1992.
N.N. Kamel, T. Song, and M. Kamel, “Incorporating GUI in integration of molecular biology databases,”Third IEEE Workshop Future Trends Distributed Computing Systems in the 1990's, 1992.
W. Litwin and A. Abdellatif, “Multi-database interoperability,”IEEE Comput., vol. 19, no. 12, 1986.
W. Litwin, L. Mark, and N. Roussopoulos, “Interoperability of multiple autonomous databases,”ACM Comput. Surveys, vol. 22, no. 3, 1986.
Open Software Foundation, OSF/Motif R 1.1 series, Prentice Hall, 1991.
R.W. Scheifler and J. Gettys, “The X window system,”ACM Trans. Graph. vol. 5, no. 2, 1986.
A.P. Sheth and J.A. Larson, “Federated database systems for managing distributed, heterogeneous, and autonomous databases,”ACM Comput. Surveys, vol. 22, no. 3, 1990.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kamel, N.N., Song, T. & Kamel, M. An approach for building an integrated environment for molecular biology databases. Distrib Parallel Databases 1, 303–327 (1993). https://doi.org/10.1007/BF01263335
Issue Date:
DOI: https://doi.org/10.1007/BF01263335