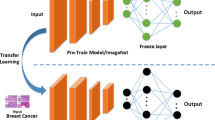
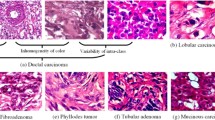
Abstract
The efficiency in breast cancer from imaging-based computer-aided diagnosis (CAD) has been revealed in recent years. As a fact, the methods grounded on a single modality constantly lack behind multimodal CAD imaging. However, owing to the restrictions of imaging devices, expressly in rural hospitals, single-modal imaging becomes a favorite in clinical practice for diagnosis. A fresh learning model trending nowadays known as learning using privileged information (LUPI) adopts additional privileged information (PI) modality to help during the training stage, but PI does not contribute in the testing stage. Meanwhile, the link exists between PI and training samples; the same is then reassigned to the learned model. We propose a LUPI-based CAD framework for breast cancer using privileged information in this work. The work offers both a classifier- or feature-level LUPI, in which the information is shifted from the additional PI modality to the diagnosis modality. A thorough comparison has been made among six classifier-level algorithms and six feature-level LUPI algorithms. The experimental results on both the acquired primary datasets show that all classifier-level and deep learning-based feature-level LUPI algorithms can enhance the performance of a single-modal imaging-based CAD for breast cancer by relocating PI.








Similar content being viewed by others
Change history
27 February 2020
The articles listed below were published in Issue January 2020, Issue 1, instead of Issue February 2020, Issues 1–2.
References
Sree, S.V., Ng, E.Y., Acharya, R.U., Faust, O.: Breast imaging: a survey. World J. Clin. Oncol. 2(4), 171–178 (2011). https://doi.org/10.5306/wjco.v2.i4.171
Soltani, M., Rahpeima, R., Kashkooli, F.M.: Breast cancer diagnosis with a microwave thermoacoustic imaging technique-a numerical approach. Med. Biol. Eng. Comput. 57, 1497–1513 (2019)
Fan, L., Strasserweippl, K., Li, J., Louis, J.: Breast cancer in China. Lancet Oncol. 15(7), 279–289 (2014)
Sizilio, G.R., Leite, C.R., Guerreiro, A.M., Neto, A.D.: Fuzzy method for pre-diagnosis of breast cancer from the fine needle aspirate analysis. Biomed. Eng. Online (2012). https://doi.org/10.1186/1475-925x-11-83
Tang, J., Rangayyan, R.M., Naqa, I.E., Yang, Y.: Computer-aided detection and diagnosis of breast cancer with mammography, Recent Advances. IEEE Trans. ITB 13(2), 236–251 (2009)
Kadam, V.J., Jadhav, S.M., Vijayakumar, K.: Breast cancer diagnosis using feature ensemble learning based on stacked sparse autoencoders and softmax regression. J. Med. Syst. 43(8), 1–11 (2019)
Abdar, M., Moghadam, M.Z., Zhou, X., Gururajan, R., Tao, X., Barua, P.D., Gururajan, R.: A new nested ensemble technique for automated diagnosis of breast cancer. Pattern Recognit. Lett. 18, 1–11 (2018)
Karabatak, M., et al.: A new classifier for breast cancer detection based on Naive Bayesian. Measurement 72, 32–36 (2015)
Diz, J., Marreiros, G., Freitas, A.: Applying data mining techniques to improve breast cancer diagnosis. J. Med. Syst. 40(203), 1–7 (2016)
Venkatesh, S.S., et al.: Going beyond a first reader: a machine learning methodology for optimizing cost and performance in breast ultrasound diagnosis. Ultrasound Med. Biol. 41(12), 3148–3162 (2015)
Bhardwaj, A., Tiwari, A.: Breast cancer diagnosis using genetically optimized neural network model. Expert Syst. Appl. 42(10), 4611–4620 (2015)
Karabatak, M., Ince, M.C.: An expert system for detection of breast cancer based on association rules and neural network. Expert Syst. Appl. 36, 3465–3469 (2009)
Azar, A.T., El-Said, S.A.: Probabilistic neural network for breast cancer classification. Neural Comput. Appl. 23(6), 1737–1751 (2013)
Chen, H., Yang, B., Liu, J., Liu, D.Y.: A support vector machine classifier with rough set-based feature selection for breast cancer diagnosis. Expert Syst. Appl. 38(7), 9014–9022 (2011)
Turki, T., Wei, Z.: Boosting support vector machines for cancer discrimination tasks. Comput. Biol. Med. 101, 236–249 (2018)
Maglogiannis, I., Zafiropoulos, E., Anagnostopoulos, I.: An intelligent system for automated breast cancer diagnosis and prognosis using SVM based classifiers. Appl. Intell. 30(1), 24–36 (2009)
Wang, H., Zheng, B., Yoon, S., Ko, H.: A support vector machine-based ensemble algorithm for breast cancer diagnosis. Eur. J. Oper. Res. 267(2), 687–699 (2018)
Basavanhally, A., Ganesan, S., Shih, N., Mies, C., Feldman, M., Tomaszewski, J., Madabhushi, A.: A boosted classifier for integrating multiple fields of view: breast cancer grading in histopathology. In: Proceedings of IEEE International Symposium on Biomedical Imaging: From Nano To Macro, pp. 125–128. Chicago (2011)
Bashir, S., Qamar, U., Khan, F.H.: Heterogeneous classifiers fusion for dynamic breast cancer diagnosis using weighted vote based ensemble. Qual. Quant. 49, 2061–2076 (2015)
Zheng, B., Yoon, S.W., Lam, S.S.: Breast cancer diagnosis based on feature extraction using a hybrid of K-means and support vector machine algorithms. Expert Syst. Appl. 41(4), 1476–1482 (2014)
Vivona, L., Cascio, D., Fauci, F., Raso, G.: Fuzzy technique for microcalcifications clustering in digital mammograms. BMC Med. Imaging 14(1), 14–23 (2014)
Lo, C.-M., et al.: Quantitative breast mass classification based on the integration of B-mode features and strain features in elastography. Comput. Biol. Med. 64, 91–100 (2015)
Onan, A., et al.: A fuzzy-rough nearest neighbor classifier combined with consistency-based subset evaluation and instance selection for automated diagnosis of breast cancer. Expert Syst. Appl. 42(20), 6844–6852 (2015)
Kaya, Y., et al.: A new intelligent classifier for breast cancer diagnosis based on rough set and extreme learning machine: RS + ELM. Turk. J. Electr. Eng. Comput. Sci. 21, 2079–2091 (2014)
Sheikhpour, R., Sarram, M.A., Sheikhpour, R.: Particle swarm optimization for bandwidth determination and feature selection of kernel density estimation based classifiers in diagnosis of breast cancer, Appl. Soft Comput. 40:113–131 (2016). arXiv:1711
Chen, H., Yan, B., Wang, J.S., Wang, G., Li, Z.H., Liu, W.: Towards an optimal support vector machine classifier, using a parallel particle swarm optimization strategy. Appl. Math. Comput. 239, 180–197 (2014)
Deng, W., Ya, R., Zhao, H., Yang, X., Li, G.: A novel intelligent diagnosis method using optimal LS-SVM with improved PSO algorithm. Soft Comput. Fusion Found. Methodol. Appl. 23(7), 2445–2462 (2019)
Peng, L., Chen, W., Zhou, W., Li, F., Yang, J., Zhang, J.: An immune-inspired semi-supervised algorithm for breast cancer diagnosis. Comput. Methods Programs Biomed. 134, 259–265 (2016)
Bhuiyan, M.N.Q., Shamsujjoha, M., Ripon, S.H., Proma, F.H., Khan, F.: Transfer learning and supervised classifier based prediction model for breast cancer. In: Dey, N., Das H., Naik, B., Behera, H.S. (eds.) Advances in Ubiquitous Sensing Applications for Healthcare, Big Data Analytics for Intelligent Healthcare Management, pp. 59–86. Academic Press, Cambridge, ISSN: 9780128181461 (2019)
Sun, W., Tseng, T.B., Zhang, J., Qian, W.: Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput. Med. Imaging Graph. 57, 4–9 (2017)
Karahaliou, A.N., Boniatis, I.S., Skiadopoulos, S.G., Sakellaropoulos, F.N., Arikidis, N.S., Likaki, E.A., Panayiotakis, G.S., Costaridou, L.I.: Breast cancer diagnosis: analyzing texture of tissue surrounding microcalcifications. IEEE Trans. Inf. Technol. Biomed. 12, 731–738 (2008). https://doi.org/10.1109/TITB.2008.920634
Andreadis, I.I., Spyrou, G.M., Nikita, K.S.: A CAD Scheme for mammography empowered with topological information from clustered microcalcifications Atlases. IEEE J. Biomed. Health Inform. 19, 166–173 (2015). https://doi.org/10.1109/JBHI.2014.2334491
Wang, J., Yang, X., Cai, H., Tan, W., Jin, C., Li, L.: Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci. Rep. 6, 27327 (2016). https://doi.org/10.1038/srep27327
Chougrad, H., Zouaki, H., Alheyane, O.: Multi-label transfer learning for the early diagnosis of breast cancer. Neurocomputing (2019). https://doi.org/10.1016/j.neucom.2019.01.112
Zhang, M.L., Zhou, Z.H.: A review on multi-label learning algorithms. IEEE Trans. Knowl. Data Eng. 26, 1819–1837 (2014)
Chougrad, H., Zouaki, H., Alheyane, O.: Convolutional neural networks for breast cancer screening: transfer learning with exponential decay. In: Proceedings of the NIPS-Machine Learning for Health Workshop (ML4H), Long Beach Convention (2017). arXiv:10752.3
Bressan, S.R., Bugatti, P.H., Saito, T.M.P.: Breast cancer diagnosis through active learning in content-based image retrieval. Neurocomputing 357, 1–10 (2019)
Shen, R., Yan, K., Tian, K., Jiang, C., Zhou, K.: Breast mass detection from the digitized X-ray mammograms based on the combination of deep active learning and self-paced learning. Future Gener. Comput. Syst. 101, 668–679 (2019)
Coletta, T.M.L., Ponti, M., Hruschka, E.R., Acharya, A., Ghosh, J.: Combining clustering and active learning for the detection and learning of new image classes. Neurocomputing 358, 150–165 (2019)
Gu, D., Liang, C., Zhao, H.: A case-based reasoning system based on weighted heterogeneous value distance metric for breast cancer diagnosis. Artif. Intell. Med. 77, 31–47 (2017)
Ahn, H., Kim, K.: Global optimization of case-based reasoning for breast cytology diagnosis. Expert Syst. Appl. 36(1), 724–734 (2009)
Krawczyk, B., Schaefer, G., Woźniak, M.: A hybrid cost-sensitive ensemble for imbalanced breast thermogram classification. Artif. Intell. Med. 65(3), 219–227 (2015)
Xu, Y., Wang, Y., Yuan, J., Cheng, Q., Wang, X., Carson, P.L.: Medical breast ultrasound image segmentation by machine learning. Ultrasonics 91, 1–9 (2019)
Fang, Z., Zhang, W., Ma, H.: Breast Cancer Classification with Ultrasound Images Based on SLIC, 1-12
Kirimasthong, K., Rodtook, A., Lohitvisate, W., Makhanov, S.S.: Automatic initialization of active contours in ultrasound images of breast cancer. Pattern Anal. Appl. 21, 491–500 (2018)
Mohammed, M.A., Al-Khateeb, B., Rashid, A.N., Ibrahim, D.A., Ghani, M.K.A., Mostafa, S.A.: Neural network and multi-fractal dimension features for breast cancer classification from ultrasound images. Comput. Electr. Eng. 000, 1–12 (2018)
Klimonda, Z., Piotrzkowska-Wroblewska, K.D.H., Karwat, P., Litniewski, J.: Quantitative ultrasound of tumor surrounding tissue for enhancement of breast cancer diagnosis. In: Proceedings 6th International Work-Conference on Bioinformatics and Biomedical Engineering (IWBBIO), pp. 186–197. Granada, Spain (2018)
Wang, N., Bian, C., Wang, Y., Xu, M., Qin, C., Yang, X., Wang, T., Li, A., Shen, D., Ni, D.: Densely deep supervised networks with threshold loss for cancer detection in automated breast ultrasound. In: Proceedings of 21st International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 641–648. Granada, Spain, 16–20 September 2018
Cheng, H.D., Shan, J., Ju, W., Guo, Y., Zhang, L.: Automated breast cancer detection and classification using ultrasound images: a survey. Pattern Recognit. 43(1), 299–317 (2010)
Salama, M.S., Eltrass, A.S., Elkamchouchi, H.M.: An improved approach for computer-aided diagnosis of breast cancer in digital mammography, published. In: IEEE International Symposium on Medical Measurements and Applications (MeMeA), Rome (2018)
Shaikh, T.A., Ali, R.: Combating breast cancer by an intelligent ensemble classifier approach. In: Proceedings of IEEE International Conference on Bioinformatics and Systems Biology (BSB), pp. 5-10. IIIT Allahabad India, October (2018)
Li, H., Meng, X., Wang, T., Tang, Y., Yin, Y.: Breast masses in mammography classification with local contour features, Biomed. Eng. Online. 16(44): 1–12, 44 (2017). https://doi.org/10.1186/s12938- 017- 0332- 0
Yassin, N.I.R., Omran, S., El Houby, E.M.F., Allam, H.: Machine learning techniques for breast cancer computer-aided diagnosis using different image modalities: a systematic review. Comput. Methods Programs Biomed. 156, 25–45 (2018)
Dhungel, N., Carneiro, G., Bradley, A.P.: A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med. Image Anal. 37, 114–128 (2017)
Tsochatzidis, L., Costaridou, L., Pratikakis, I.: Deep learning for breast cancer diagnosis from mammograms—a comparative study. J. Imaging MPDI 5(37), 1–11 (2019). https://doi.org/10.3390/jimaging5030037
Kelly, K.M., Dean, J., Comulada, W.S., Lee, S.J.: Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur. Radiol. 20(3), 734–742 (2010)
Golatta, M., Franz, D., Harcos, A.: Interobserver reliability of automated breast volume scanner (ABVS) interpretation and agreement of ABVS findings with handheld breast ultrasound (HHUS), mammography and pathology results. Eur. J. Radiol. 82(8), 332–336 (2013)
Wilczek, B., Wilczek, H.E., Rasouliyan, L., Leifland, K.: Adding 3D automated breast ultrasound to mammography screening in women with heterogeneously and extremely dense breasts: report from a hospital-based, high-volume, single-center breast cancer screening program. Eur. J. Radiol. 85(9), 1554–1563 (2016)
Cong, J., Wei, B., He, Y., Yin, Y., Zheng, Y.: A selective ensemble classification method combining mammography images with ultrasound images for breast cancer diagnosis. Hindawi Math. Methods Med. 2017, 4896386 (2017)
Yang, S.N., Li, F.J., Liao, Y.H., Chen, Y.S., Shen, W.C., Huang, T.C.: Identification of breast cancer using integrated information from MRI and mammography. PLoS ONE (2015). https://doi.org/10.1371/journal.pone.0128404
Geisel, J., Raghu, M., Hooley, R.: The role US in breast cancer screening: the case for and against ultrasound. In: Seminars in Ultrasound, CT, and MRI, vol. 39, no. 1, pp. 25–34 (2018). http://dx.doi.org/10.1053/j.sult.2017.09.006
Barth, V., et al.: Diagnosis of Breast Diseases: Integrating the Findings of Clinical Presentation, Mammography, and Ultrasound, 1st Edn. Thieme, New York, ISBN-13:978-3131438317 (2012)
Health Quality Ontario, Ultrasound as an Adjunct to Mammography for Breast Cancer Screening: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 16 (5):1–71 (2016). http://www.hqontario.ca/Evidence-to-Improve-Care/Journal-Ontario-Health-Technology-Assessment-Series. Accessed 3 March 2019
Vapnik, V., et al.: Estimation of Dependencies Based on Empirical Data, Empirical Inference Science, 2nd edn. Springer, New York (2006). ISBN 978-0-387-34239-9
Liang, L., Cherkassky, V.: Connection between SVM and multi-task learning. In: Proceedings of IEEE International Joint Conference on Neural Networks (INNS), pp. 2048–2054. Hong Kong, (2008)
Li, Y., Meng, F., Shi, J., Alzheimer’s Disease Neuroimaging Initiative: Learning using privileged information improves neuroimaging-based CAD of Alzheimer’s disease: a comparative study. Med. Biol. Eng. Comput. 57(7), 1605–1616 (2019)
Hou, Q., Zhen, L., Deng, N., Jing, L.: Novel Grouping Method-based support vector machine plus for structured data. Neurocomputing 211, 191–201 (2016)
Lapin, M., Hein, M., Schiele, B.: Learning using privileged information: SVM + and weighted SVM. Neural Netw. 53, 95–108 (2014)
Frank,A., Asuncion, A.: UCI machine learning repository. http://archive.ics.uci.edu/ml (2010). Accessed 10 Feb 2019
Vapnik, V., Vashist, A.: A new learning paradigm: learning using privileged information. Neural Netw. 22, 544–557 (2009)
Zhu, W., Zhong, P.: A new one-class SVM based on hidden information. Knowl. Based Syst. 60, 35–43 (2014)
Shi, J., Xue, Z., Dai, Y., Peng, B., Dong, Y., Zhang, Q., Zhang, Y.C.: Cascaded multi-column RVFL + classifier for single-modal neuroimaging—based diagnosis of Parkinson’s disease. IEEE Trans. Biomed. Eng. 66(8), 2362–2371 (2019)
Yu, X., Zeng, N., Liu, S., Zhang, Y.D.: Utilization of DenseNet201 for diagnosis of breast abnormality. Mach. Vis. Appl. (2019). https://doi.org/10.1007/s00138-019-01042-8
Zhang, Y.D., Pan, C., Chen, X., Wang, F.: Abnormal breast identification by nine-layer convolutional neural network with parametric rectified linear unit and rank-based stochastic pooling. J. Comput. Sci. 27, 57–68 (2018)
Shen, L., Margolies, L.R., Rothstein, J.H., Fluder, E., McBride, R., Sieh, W.: Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 9, 12495 (2019). https://doi.org/10.1038/s41598-019-48995-4
Rouhi, R., Jafari, M., Kasaei, S., Keshavarzian, P.: Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst. Appl. 42, 990–1002 (2015)
Ting, F.F., Tan, Y.J., Sim, K.S.: Convolutional neural network improvement for breast cancer classification. Expert Syst. Appl. 120, 103–115 (2019)
Tsochatzidis, L., Costaridou, L., Pratikakis, I.: Deep learning for breast cancer diagnosis from mammograms—a comparative study. J. Imaging MPDI 5, 37 (2019)
Huang, H., Shi, L., Suykens, J.A.K.: Support vector machine classifier with pinball loss. IEEE Trans. Pattern Anal. Mach. Intell. 36(5), 984–997 (2014)
Zhu, W., et al.: A new support vector machine plus with pinball loss. J. Classif. 35(1), 52–70 (2018)
Zhang P.B., Yang, Z.X.: A new learning paradigm for the random vector functional-link network: RVFL + (2019) arXiv:1708.08282
Rastegar, S., Baghshah, M.S., Rabiee, H.R., Shojaee, S.M.: MDLCW: a multimodal deep learning framework with cross weights. In: Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (CVPR), pp. 2601–2609. Salt lake city (2016)
Srivastava, N., Salakhutdinov, R.: Multimodal learning with deep boltzmann machines. J. Mach. Learn. Res. 15, 2949–2980 (2014)
Wang, W., Arora, R., Livescu, K., Bilmes, J.A.: Unsupervised learning of acoustic features via deep canonical correlation analysis. In Proceedings of IEEE International Conference on Acoustics, Speech and Signal Processing (CASSP), pp. 4590–4594. (2015)
Sohn, K., Shang, W., Lee, H.: Improved multimodal deep learning with the variation of information. In Proceedings of 27th International Conference on Neural Information Processing Systems (NIPS’14), pp. 2141–2149. Montreal (2014)
Andrew, G., Arora, R., Bilmes, A., Livescu, K.: Deep canonical correlation analysis. In Proceedings of International Conference on Machine Learning (ICML), pp. 1247–1255. Atlanta (2013)
Shrivastava, A., Chaudhary, A., Kulshreshtha, D., Prakash Singh, V., Srivastava, R.: Automated digital mammogram segmentation using dispersed region growing and sliding window algorithm. In: Proceedings of 2nd International Conference on Image, Vision, and Computing, pp. 366–370. Chengdu (2017)
Chan, H.P., Vyborny, C.J., MacMahon, H.E., Metz, C.E., Doi, K., Sickles, E.A.: Digital mammography: ROC studies of the effects of pixel size and unsharp-mask filtering on the detection of subtle microcalcifications. Investig. Radiol. 22(7), 581–589 (1987)
Vincent, L., Soille, P.: Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 13(6), 583–593 (1991)
Reyad, Y.A., Berbar, M.A., Hussain, M.: Comparison of statistical LBP and multi-resolution analysis features for breast mass classification. J. Med. Syst. 38(9), 1–15 (2014)
Alahmadi, H.H., Shen, Y., Fouad, S., Luft, C.D.B., Bentham, P., Kourtzim, Z., Tino, L.P.: Classifying cognitive profiles using machine learning with privileged information in mild cognitive impairment. Front. Comput. Neurosci. 10(117), 1–17 (2016)
Gao, Z., Wu, S., Liu, Z., Luo, J., Zhang, H., Gong, M., Li, S.: Learning the implicit strain reconstruction in ultrasound elastography using privileged information. Med. Image Anal. 58(101534), 1–16 (2019)
Gautam, C., Tiwari, A., Tanveer, M.: KOC +: kernel ridge regression-based one-class classification using privileged information. Inf. Sci. 504, 324–333 (2019)
Li, Y., Meng, F., Shi, J.: and Alzheimer’s disease neuroimaging initiative, learning using privileged information improves neuroimaging-based CAD of Alzheimer’s disease: a comparative study. Med. Biol. Eng. Comput. 57, 1605–1616 (2019)
Shi, J., Xue, Z., Dai, Y., Peng, B., Dong, Y., Zhang, Q., Zhang, Y.: Cascaded multi-column RVFL + classifier for single-modal neuroimaging-based diagnosis of Parkinson’s disease. IEEE Trans. Biomed. Eng. 24, 1–10 (2018)
Li, W., Dai, D., Tan, M., Xu, D., Gool, L.V.: Fast algorithms for linear and kernel SVM + . In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2258–2266. (2016)
Acknowledgements
Authors are very much thankful to doctors/radiologists and staff team of the Department of Radiology, Jawaharlal Nehru Medical College (JNMC), Aligarh Muslim University, India and medical imaging centers, i.e., Medicare Diagnostic Center and DM diagnostic clinic, Srinagar, India, for verifying all the tissue segmentation results carefully and rating the results on the scale.
Funding
This work is partly supported by the Research Fellowship of the ‘Visvesvaraya Ph.D. Scheme for Electronics & IT,’ Ministry of Electronics and Information technology (MeitY), Government of India (GoI), Vide Grant no. PhD-MLA/4(39)/2015-16.
Author information
Authors and Affiliations
Contributions
Mr. TAS conceived, designed, and performed the experiments. Mr. RA analyzed the data, and he offered his valuable suggestions and feedbacks. Both contributed to reagents/materials/analysis tools and paper writing jointly.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shaikh, T.A., Ali, R. & Beg, M.M.S. Transfer learning privileged information fuels CAD diagnosis of breast cancer. Machine Vision and Applications 31, 9 (2020). https://doi.org/10.1007/s00138-020-01058-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00138-020-01058-5