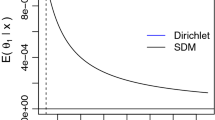
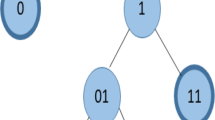
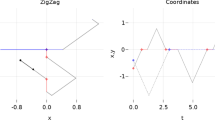
Abstract
Sparse Markov models (SMMs) provide a parsimonious representation for higher-order Markov models. We present a computationally efficient method for fitting SMMs using a collapsed Gibbs sampler, the GSDPMM. We prove the consistency of the GSDPMM in fitting SMMs. In simulations, the GSDPMM was found to perform as well or better than existing methods for fitting SMMs. We apply the GSDPMM method to fit SMMs to patterns of wind speeds and DNA sequences.

Similar content being viewed by others
References
Aitchison J, Barceló-Vidal C, Martán-Ferníandez JA, Pawlowsky-Glahn V (2000) Logratio analysis and compositional distance. Math Geol 32:271–275
Almagor H (1983) A Markov analysis of DNA sequences. J Theor Biol 104(4):633–645. https://doi.org/10.1016/0022-5193(83)90251-5
Avery P (1987) The analysis of intron data and their use in the detection of short signals. J Mol Evol 26:335–340
Blei DM, Jordan MI (2006) Variational inference for Dirichlet process mixtures. Bayesian Anal 1(1):121–143
Borges J, Levene M (2007) Evaluating variable-length Markov chain models for analysis of user web navigation sessions. IEEE Trans Knowl Data Eng 19(4):441–452
Bühlmann P, Wyner AJ (1999) Variable length Markov chains. Ann Stat 27(2):480–513. https://doi.org/10.1214/aos/1018031204
Dai Q, Liu X-Q, Wang T-M (2006) Numerical characterization of DNA sequences based on the k-step Markov chain transition probability. J Comput Chem 27(15):1830–1842
Ferguson TS (1973) A Bayesian analysis of some nonparametric problems. Ann Stat 1(2):209–230. https://doi.org/10.1214/aos/1176342360
Garcia J, Gonzalez-Lopez V (2017) Consistent estimation of partition Markov models. Entropy. https://doi.org/10.3390/e19040160
Garcia J, Gonzalez-Lopez V (2010) Minimal Markov models. arXiv:1002.0729 [math.ST]
Görür D, Rasmussen CE (2010) Dirichlet process Gaussian mixture models: choice of the base distribution. J Comput Sci Technol 25(4):653–664
Haslett J, Raftery AE (1989) Space-time modelling with long-memory dependence: assessing Ireland’s wind power resource. J R Stat Soc Ser C (Appl Stat) 38(1):1–50
Hubert L, Arabie P (1985) Comparing partitions. J Classif 2(1):193–218
Jääskinen V, Xiong J, Corander J, Koski T (2014) Sparse Markov chains for sequence data. Scand J Stat 41(3):639–655. https://doi.org/10.1111/sjos.12053
Kharin Y (2017) Statistical analysis of big data based on parsimonious models of high-order Markov chains, pp 485–496. https://doi.org/10.1007/978-3-319-71504-940
Kharin Y, Petlitskii AI (2007) A Markov chain of order s with r partial connections and statistical inference on its parameters. Discrete Math Appl 17(3):295–317. https://doi.org/10.1515/dma.2007.026
Kullback S, Leibler RA (1951) On information and sufficiency. Ann Math Stat 22(1):79–86. https://doi.org/10.1214/aoms/1177729694
Neal RM (2000) Markov chain sampling methods for Dirichlet process mixture models. J Comput Graph Stat 9(2):249–265
Pian C, Yang Z, Yang Y, Zhang L, Chen Y (2021) Identifying RNA N6-methyladenine sites in three species based on a Markov model. Front Genet. https://doi.org/10.3389/fgene.2021.650803
Ptaszynski M, Momouchi Y (2012) Part-of-speech tagger for Ainu language based on higher order hidden Markov model. Expert Syst Appl 39(14):11576–11582. https://doi.org/10.1016/j.eswa.2012.04.031
Raftery A, Tavare S (1994) Estimation and modelling repeated patterns in high order Markov chains with the mixture transition distribution model. J R Stat Soc Ser C (Appl Stat) 43(1):179–199
Sanjari MJ, Gooi HB (2017) Probabilistic forecast of PV power generation based on higher order Markov chain. IEEE Trans Power Syst 32(4):2942–2952
Sarkar A, Dunson DB (2016) Bayesian nonparametric modeling of higher order Markov chains. J Am Stat Assoc 111(516):1791–1803
Xiong J, Jääskinen V, Corander J (2016) Recursive learning for sparse Markov models. Bayesian Anal 11(1):247–263. https://doi.org/10.1214/15-BA949
Yang J, Lang K, Zhang G, Fan X, Chen Y, Pian C (2020) SOMM4mC: a secondorder Markov model for DNA N4-methylcytosine site prediction in six species. Bioinformatics 36(14):4103–4105. https://doi.org/10.1093/bioinformatics/btaa507
Yin J, Wang J (2016) A model-based approach for text clustering with outlier detection. 2016 IEEE 32nd Int Conf Data Eng (ICDE) 625–636
Zhang J, Ghahramani Z, Yang Y (2005) A probabilistic model for online document clustering with application to novelty detection. In: Saul LK, Weiss Y, Bottou L (eds) Advances in neural information processing systems 17. MIT Press, pp 1617–1624
Zhu D-M, Lu J, Ching W-K, Siu T-K (2017) Discrete-time optimal asset allocation under higher-order hidden Markov model. Econ Model 66:223–232. https://doi.org/10.1016/j.econmod.2017.07.00628
Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant No. 1811933.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bennett, I., Martin, D.E.K. & Lahiri, S.N. Fitting sparse Markov models through a collapsed Gibbs sampler. Comput Stat 38, 1977–1994 (2023). https://doi.org/10.1007/s00180-022-01310-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-022-01310-8