Abstract
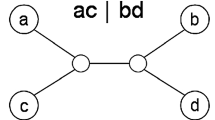
We present a new distance based quartet method for phylogenetic tree reconstruction, called Minimum Tree Cost Quartet Puzzling. Starting from a distance matrix computed from natural data, the algorithm incrementally constructs a tree by adding one taxon at a time to the intermediary tree using a cost function based on the relaxed 4-point condition for weighting quartets. Different input orders of taxa lead to trees having distinct topologies which can be evaluated using a maximum likelihood or weighted least squares optimality criterion. Using reduced sets of quartets and a simple heuristic tree search strategy we obtain an overall complexity of O(n 5 log2 n) for the algorithm. We evaluate the performances of the method through comparative tests and show that our method outperforms NJ when a weighted least squares optimality criterion is employed. We also discuss the theoretical boundaries of the algorithm.
Similar content being viewed by others
References
BANDELT, H., and DRESS, A. (1992), “Split Decomposition: A New and Useful Approach to Phylogenetic Analysis of Distance Data”, Molecular Phylogenetics and Evolution, 1, 242–252.
BANDELT, H. J., and DRESS, A. (1986), “Reconstructing the Shape of a Tree from Observed Dissimilarity Data”, Advances in Applied Mathematics, 7, 309–343.
BERRY, V., and GASCUEL, O. (2000), “Inferring Evolutionary Trees with Strong Combinatorial Evidence”, Theoretical Computer Science, 240, 271–298.
BERRY, V., JIANG, T., KEARNEY, P., LI, M., and WAREHAM, T. (1999), “Quartet Cleaning: Improved Algorithms and Simulations”, in ESA ’99: Proceedings of the 7th Annual European Symposium on Algorithms, London UK: Springer-Verlag, pp. 313–324.
BUNEMAN, P. (1971), “The Recovery of Trees from Measures of Dissimilarity”, Mathematics in the Archaeological and Historical Sciences, 387–395.
CILIBRASI, R., and VITANYI, P. (2005), “A New Quartet Tree Heuristic for Hierarchical Clustering”, in Principled Methods of Trading Exploration and Exploitation Workshop, London UK.
ERD ÖS, P., STEEL, M., SZ´EKELY, L. A., and WARNOW, T. (1997), “Constructing Big Trees from Short Sequences”, in Automata, Languages and Programming, Lecture Notes in Computer Science, Vol. 1256, Heidelberg, Germany: Springer-Verlag, pp. 827–837.
FELSENSTEIN, J. (1981), “Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach”, Molecular Biology and Evolution, 17, 368–376.
FELSENSTEIN, J. (1997), “An Alternating Least Squares Approach to Inferring Phylogenies from Pairwise Distances”, Systematic Biology, 46, 101–111.
FELSENSTEIN, J. (2005), PHYLIP (Phylogeny Inference Package) Version 3.6, distributed by the author, Department of Genome Sciences, University of Washington, Seattle.
FITCH,W. (1981), “A Non-sequential Method for Constructing Trees and Hierarchical Classifications”, Molecular Biology and Evolution, 18, 30–37.
GASCUEL, O. (1997), “BIONJ: An Improved Version of the NJ Algorithm Based on a Simple Model of Sequence Data”, Molecular Biology and Evolution, 14, 685–695.
GRAMM, J., and NIEDERMEIER, R. (2003), “A Fixed-parameter Algorithm for Minimum Quartet Inconsistency”, Journal of Computer and System Sciences, 67, 723–741.
HOLLAND, B., HUBER, K., DRESS, A., and MOULTON, V. (2002), “δ Plots: A Tool for Analyzing Phylogenetic Distance Data”, Molecular Biology and Evolution, 19, 2051–2059.
HUSON, D., SMITH, K., and WARNOW, T. (1999), “Estimating Large Distances in Phylogenetic Reconstruction”, in Algorithm Engineering, eds. J. Vitter and C. Zaroliagis, Lecture Notes in Computer Science, Vol. 1668, London UK: Springer-Verlag, pp. 271–285.
JIANG, T., KEARNEY, P., and LI, M. (1998), “Orchestrating Quartets: Approximation and Data Correction”, in FOCS ’98: Proceedings of the 39th Annual Symposium on Foundations of Computer Science, IEEE Computer Society, Washington DC, USA, p. 416.
JUKES, T. H., and CANTOR, C. R. (1969), Evolution of Protein Molecules, NewYork: Academy Press.
KEARNEY, P. (1998), “The Ordinal Quartet Method”, in RECOMB ’98: Proceedings of the Second Annual International Conference on Computational molecular biology, ACM, New York, NY, USA, pp. 125–134.
MARGUSH, T., and MCMORRIS, F. R. (1981), “Consensus n-Trees”, Bulletin of Mathematical Biology, 42, 239–244.
RAMBAUT, A., and GRASS, N. (1997), “Seq-Gen: An Application for the Monte Carlo Simulation of DNA Sequence Evolution Along Phylogenetic Trees”, Computer Applications in the Biosciences, 13, 235–238.
RANWEZ, V., and GASCUEL, O. (2001), “Quartet Based Phylogenetic Inference: Improvements and Limits”, Molecular Biology and Evolution, 18, 1103–1116.
ROBINSON, D., and FOULDS, L. (1981), “Comparison of Phylogenetic Trees”, Mathematical Biosciences, 53, 131–147.
SAITOU, N., and NEI, M. (1987), “The Neighbor-joining Method: A New Method for Reconstructing Phylogenetic Trees”, Molecular Biology and Evolution, 4, 406–25.
SATTATH, S., and TVERSKY, A. (1977), “Additive Similarity Trees”, Psychometrika, 42, 319–345.
SCHMIDT, H., STRIMMER, K., VINGRON, M., and VON HAESELER, A. (2002), “TREE-PUZZLE:Maximum Likelihood Phylogenetic Analysis Using Quartets and Parallel Computing”, Bioinformatics, 18, 502–504.
SFORZA, C., and EDWARDS, A. (1967), “Phylogenetic Analysis: Models and Estimation Procedures”, American Journal of Human Genetics, 19, 223–257.
SNIR, S., WARNOW, T., and RAO, S. (2007), “Short Quartet Puzzling: A New Quartet-Based Phylogeny Reconstruction Algorithm”, Journal of Computational Biology, 15, 91–103.
ST. JOHN, K.,WARNOW, T.,MORET, B., and VAWTER, L. (2003), “Performance Study of Phylogenetic Methods: (Unweighted) Quartet Methods and Neighbor-joining”, Journal of Algorithms, 48, 173–193.
STEEL, M. (1992), “The Complexity of Reconstructing Trees from Qualitative Characters and Subtrees”, Journal of Classification, 9, 91–116.
STRIMMER, K., and VON HAESELER, A. (1996), “Quartet Puzzling: A Quartet Maximum-Likelihood Method for Reconstructing Tree Topologies”, Molecular Biology and Evolution, 13, 964–969.
VINH, L., and VON HAESELER, A. (2004), “IQPNNI: Moving Fast Through Tree Space and Stopping in Time”, Molecular Biology and Evolution, 21, 1565–1571.
WU, G., YOU, J., and LIN, G. (2006), “A Polynomial Algorithm for the Minimum Quartet Inconsistency Problem with O(n) Quartet Errors”, Information Processing Letters, 100, 167–171.
Author information
Authors and Affiliations
Corresponding author
Additional information
We would like to thank the editor and two anonymous referees for useful comments on earlier versions of this paper.
Rights and permissions
About this article
Cite this article
Ionescu, T.B., Polaillon, G. & Boulanger, F. Minimum Tree Cost Quartet Puzzling. J Classif 27, 136–157 (2010). https://doi.org/10.1007/s00357-010-9053-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00357-010-9053-9