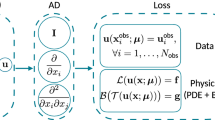
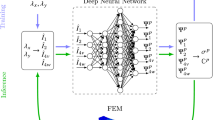
Abstract
Many natural materials exhibit highly complex, nonlinear, anisotropic, and heterogeneous mechanical properties. Recently, it has been demonstrated that data-driven strain energy functions possess the flexibility to capture the behavior of these complex materials with high accuracy while satisfying physics-based constraints. However, most of these approaches disregard the uncertainty in the estimates and the spatial heterogeneity of these materials. In this work, we leverage recent advances in generative models to address these issues. We use as building block neural ordinary equations (NODE) that—by construction—create polyconvex strain energy functions, a key property of realistic hyperelastic material models. We combine this approach with probabilistic diffusion models to generate new samples of strain energy functions. This technique allows us to sample a vector of Gaussian white noise and translate it to NODE parameters thereby representing plausible strain energy functions. We extend our approach to spatially correlated diffusion resulting in heterogeneous material properties for arbitrary geometries. We extensively test our method with synthetic and experimental data on biological tissues and run finite element simulations with various degrees of spatial heterogeneity. We believe this approach is a major step forward including uncertainty in predictive, data-driven models of hyperelasticity.











Similar content being viewed by others
Data availability
All data, model parameters, and code associated with this study are available in a publich repository at https://github.com/tajtac/node_diffusion
References
Lanir Y (2017) Multi-scale structural modeling of soft tissues mechanics and mechanobiology. J Elast 129(1–2):7–48
Jor JW, Parker MD, Taberner AJ, Nash MP, Nielsen PM (2013) Computational and experimental characterization of skin mechanics: identifying current challenges and future directions. Wiley Interdiscip Rev Syst Biol Med 5(5):539–556
Jin H, Zhang E, Espinosa HD (2023) Recent advances and applications of machine learning in experimental solid mechanics: A review. Appl Mech Rev. https://doi.org/10.1115/1.4062966
Dal H, Denli FA, Açan AK, Kaliske M (2023). Data-Driven Hyperelasticity - Part I: A Canonical Isotropic Formulation for Rubberlike Materials. https://doi.org/10.2139/ssrn.4508297
Eghtesad A, Fuhg JN, Bouklas N (2023) NN-EVP: A physics informed neural network-based elasto-viscoplastic framework for predictions of grain size-aware flow response under large deformations. arXiv:2307.04301
Rosenkranz M, Kalina KA, Brummund J, Kästner M (2023) A comparative study on different neural network architectures to model inelasticity. Int J Numer Methods Eng. https://doi.org/10.1002/nme.7319
Sacks MS, Motiwale S, Goodbrake C, Zhang W (2022) Neural Network Approaches for Soft Biological Tissue and Organ Simulations. J Biomech Eng 144(12):121010. https://doi.org/10.1115/1.4055835
Liu M, Liang L, Sun W (2020) A generic physics-informed neural network-based constitutive model for soft biological tissues. Comput Methods Appl Mech Eng 372:5
Tac V, Sree VD, Rausch MK, Tepole AB (2022) Data-driven modeling of the mechanical behavior of anisotropic soft biological tissue. Eng Comput 38(5):4167–4182
Leng Y, Tac V, Calve S, Tepole AB (2021) Predicting the mechanical properties of biopolymer gels using neural networks trained on discrete fiber network data. Comput Methods Appl Mech Eng 387:114160. https://doi.org/10.1016/j.cma.2021.114160
Kalina KA, Linden L, Brummund J, Metsch P, Kästner M (2022) Automated constitutive modeling of isotropic hyperelasticity based on artificial neural networks. Comput Mech 69(1):213–232
Fuhg JN, Bouklas N, Jones RE (2022) Learning hyperelastic anisotropy from data via a tensor basis neural network. J Mech Phys Solids 168:105022. https://doi.org/10.1016/j.jmps.2022.105022
Aggarwal A, Jensen BS, Pant S, Lee C-H (2023) Strain energy density as a Gaussian process and its utilization in stochastic finite element analysis: Application to planar soft tissues. Comput Methods Appl Mech Eng 404:115812. https://doi.org/10.1016/j.cma.2022.115812
Klein DK, Fernández M, Martin RJ, Neff P, Weeger O (2022) Polyconvex anisotropic hyperelasticity with neural networks. J Mech Phys Solids 159:104703
Linka K, Kuhl E (2023) A new family of constitutive artificial neural networks towards automated model discovery. Comput Methods Appl Mech Eng 403:115731
Tac V, Sahli Costabal F, Tepole AB (2022) Data-driven tissue mechanics with polyconvex neural ordinary differential equations. Comput Methods Appl Mech Eng 398:115248. https://doi.org/10.1016/j.cma.2022.115248
Taç V, Linka K, Sahli-Costabal F, Kuhl E, Tepole AB (2023) Benchmarking physics-informed frameworks for data-driven hyperelasticity. Comput Mech. https://doi.org/10.1007/s00466-023-02355-2
Flaschel M, Kumar S, De Lorenzis L (2023) Automated discovery of generalized standard material models with euclid. Comput Methods Appl Mech Eng 405:115867
Thakolkaran P, Joshi A, Zheng Y, Flaschel M, De Lorenzis L, Kumar S (2022) Nn-euclid: Deep-learning hyperelasticity without stress data. J Mech Phys Solids 169:105076
Wang Z, Estrada JB, Arruda EM, Garikipati K (2021) Inference of deformation mechanisms and constitutive response of soft material surrogates of biological tissue by full-field characterization and data-driven variational system identification. J Mech Phys Solids 153:104474
St. Pierre SR, Rajasekharan D, Darwin EC, Linka K, Levenston ME, Kuhl E (2023) Discovering the mechanics of artificial and real meat (Jun. 2023). https://doi.org/10.1101/2023.06.04.543638
Liang L, Liu M, Elefteriades J, Sun W (2023) Pytorch-fea: Autograd-enabled finite element analysis methods with applications for biomechanical analysis of human aorta. Computer Methods and Programs in Biomedicine 238:107616 https://doi.org/10.1016/j.cmpb.2023.107616. URL https://www.sciencedirect.com/science/article/pii/S016926072300281X
Xue T, Liao S, Gan Z, Park C, Xie X, Liu WK, Cao J JAX-FEM: A differentiable GPU-accelerated 3D finite element solver for automatic inverse design and mechanistic data science
Klein DK, Roth FJ, Valizadeh I, Weeger O (2023) Parametrised polyconvex hyperelasticity with physics-augmented neural networks (Jul. 2023). arXiv:2307.03463
Matouš K, Geers MG, Kouznetsova VG, Gillman A (2017) A review of predictive nonlinear theories for multiscale modeling of heterogeneous materials. J Comput Phys 330:192–220
Kouznetsova V, Brekelmans W, Baaijens F (2001) An approach to micro-macro modeling of heterogeneous materials. Comput Mech 27(1):37–48
Li X, Liu Z, Cui S, Luo C, Li C, Zhuang Z (2019) Predicting the effective mechanical property of heterogeneous materials by image based modeling and deep learning. Comput Methods Appl Mech Eng 347:735–753
Lee T, Bilionis I, Tepole AB (2020) Propagation of uncertainty in the mechanical and biological response of growing tissues using multi-fidelity gaussian process regression. Comput Methods Appl Mech Eng 359:112724
Stowers C, Lee T, Bilionis I, Gosain AK, Tepole AB (2021) Improving reconstructive surgery design using gaussian process surrogates to capture material behavior uncertainty. J Mech Behav Biomed Mater 118:104340
Jolicoeur-Martineau A, Piché-Taillefer R, des Combes RT, Mitliagkas I (Oct. 2020) Adversarial score matching and improved sampling for image generation. arXiv:2009.05475
Chen N, Zhang Y, Zen H, Weiss RJ, Norouzi M, Chan W (2020) WaveGrad: Estimating Gradients for Waveform Generation (Oct. 2020). arXiv:2009.00713
Lee JS, Kim J, Kim PM (2023) Proteinsgm: Score-based generative modeling for de novo protein design. https://doi.org/10.1101/2022.07.13.499967. URL https://www.biorxiv.org/content/early/2023/02/04/2022.07.13.499967
Pidstrigach J (2022) Score-based generative models detect manifolds. arXiv:2206.01018
Song Y, Sohl-Dickstein J, Kingma DP, Kumar A, Ermon S, Poole B (Feb. 2021) Score-Based Generative Modeling through Stochastic Differential Equations. arXiv:2011.13456
Croitoru, F-A, Hondru V, Ionescu RT, Shah M (2023) Diffusion Models in Vision: A Survey, IEEE Transactions on Pattern Analysis and Machine Intelligence 1–20 arXiv:2209.04747, https://doi.org/10.1109/TPAMI.2023.3261988
Taç V, Rausch MK, Costabal FS, Tepole B (2023) Data-driven anisotropic finite viscoelasticity using neural ordinary differential equations. Comput Methods Appl Mech Eng 411:98
Karras T, Aittala M, Aila T, Laine S (2022) Elucidating the design space of diffusion-based generative models. Adv Neural Inf Process Syst 35:26565–26577
Pidstrigach J (2022) Score-based generative models introduction. URL https://jakiw.com/sgm_intro
Vincent P (2011) A connection between score matching and denoising autoencoders. Neural Comput 23(7):1661–1674
Chung H, Sim B, Ryu D, Ye JC (2022) Improving diffusion models for inverse problems using manifold constraints. Adv Neural Inf Process Syst 35:25683–25696
Chung H, Kim J, Mccann MT, Klasky ML, Ye JC (2022) Diffusion posterior sampling for general noisy inverse problems, arXiv preprint arXiv:2209.14687
Du Y, Collins K, Tenenbaum J, Sitzmann V (2021) Learning signal-agnostic manifolds of neural fields. Adv Neural Inf Process Syst 34:8320–8331
Dupont E, Kim H, Eslami S, Rezende D, Rosenbaum D (2022) From data to functa: Your data point is a function and you can treat it like one, arXiv preprint arXiv:2201.12204
Elhag AA, Susskind JM, Bautista MA (2023) Manifold diffusion fields, arXiv preprint arXiv:2305.15586
Borovitskiy V, Terenin A, Mostowsky P et al (2020) Matérn gaussian processes on riemannian manifolds. Adv Neural Inf Process Syst 33:12426–12437
Gander L, Pezzuto S, Gharaviri A, Krause R, Perdikaris P, Sahli Costabal F (2022) Fast characterization of inducible regions of atrial fibrillation models with multi-fidelity gaussian process classification. Front Physiol 260:2
Hughes TJ (2012) The finite element method: linear static and dynamic finite element analysis. Courier Corporation
May-Newman K, Yin F (1998) A constitutive law for mitral valve tissue
Meador WD, Sugerman GP, Story HM, Steifert AW, Bersi MR, Tepole AB, Rausch MK (2020) The regional-dependent biaxial behavior of young and aged mouse skin: A detailed histomechanical characterization, residual strain analysis, and constitutive model. Acta Biomater 101:403–413
Rizzo ML, Székely GJ (2016) Energy distance. Wiley Interdiscip Rev Comput Stat 8(1):27–38. https://doi.org/10.1002/wics.1375
Luebberding S, Krueger N, Kerscher M (2014) Mechanical properties of human skin in vivo: a comparative evaluation in 300 men and women. Skin Res Technol 20(2):127–135
Lee T, Turin SY, Stowers C, Gosain AK, Tepole AB (2021) Personalized computational models of tissue-rearrangement in the scalp predict the mechanical stress signature of rotation flaps. Cleft Palate Craniofac J 58(4):438–445
Krueger D, Huang C-W, Islam R, Turner R, Lacoste A, Courville A (2017) Bayesian hypernetworks, arXiv preprint arXiv:1710.04759
Yang L, Zhang Z, Song Y, Hong S, Xu R, Zhao Y, Shao Y, Zhang W, Cui B, Yang M-H (2022) Diffusion models: A comprehensive survey of methods and applications, arXiv preprint arXiv:2209.00796
Zhuang P, Abnar S, Gu J, Schwing A, Susskind JM, Bautista MÁ (2022) Diffusion probabilistic fields, in: The Eleventh International Conference on Learning Representations
Dutordoir V, Saul A, Ghahramani Z, Simpson F (2023) Neural diffusion processes, in: International Conference on Machine Learning, PMLR, pp. 8990–9012
Staber B, Guilleminot J (2018) A random field model for anisotropic strain energy functions and its application for uncertainty quantification in vascular mechanics. Comput Methods Appl Mech Eng 333:94–113
Hauseux P, Hale JS, Cotin S, Bordas SP (2018) Quantifying the uncertainty in a hyperelastic soft tissue model with stochastic parameters. Appl Math Model 62:86–102
Joodaki H, Panzer MB (2018) Skin mechanical properties and modeling: a review. J Eng Med 232:4
Lee T, Turin SY, Gosain AK, Bilionis I, Tepole AB (2018) Propagation of material behavior uncertainty in a nonlinear finite element model of reconstructive surgery. Biomech Model Mechanobiol 17:1857–1873
Mueller B, Elrod J, Distler O, Schiestl C, Mazza E (2021) On the reliability of suction measurements for skin characterization. J Biomech Eng 143(2):021002
Laiacona D, Cohen J, Coulon K, Lipsky ZW, Maiorana C, Boltyanskiy R, Dufresne ER, German GK (2019) Non-invasive in vivo quantification of human skin tension lines. Acta Biomater 88:141–148
Liang X, Boppart SA (2009) Biomechanical properties of in vivo human skin from dynamic optical coherence elastography. IEEE Trans Biomed Eng 57(4):953–959
Song G, An J, Tepole AB, Lee T (2022) Bayesian inference with gaussian process surrogates to characterize anisotropic mechanical properties of skin from suction tests. J Biomech Eng 144(12):121003
Kakaletsis S, Meador WD, Mathur M, Sugerman GP, Jazwiec T, Malinowski M, Lejeune E, Timek TA, Rausch MK (2021) Right ventricular myocardial mechanics: Multi-modal deformation, microstructure, modeling, and comparison to the left ventricle. Acta Biomater 123:154–166
Meador WD, Mathur M, Sugerman GP, Jazwiec T, Malinowski M, Bersi MR, Timek TA, Rausch MK (2020) A detailed mechanical and microstructural analysis of ovine tricuspid valve leaflets. Acta Biomater 102:100–113
Chen S, Ní Annaidh A, Roccabianca S (2020) A microstructurally inspired constitutive model for skin mechanics. Biomech Model Mechanobiol 19(1):275–289
Erickson CB, Ankenman BE, Sanchez SM (2018) Comparison of gaussian process modeling software. Eur J Oper Res 266(1):179–192
Costabal FS, Pezzuto S, Perdikaris P (2022) Delta -pinns: physics-informed neural networks on complex geometries, arXiv preprint arXiv:2209.03984
You H, Zhang Q, Ross CJ, Lee C-H, Hsu M-C, Yu Y (2022) A physics-guided neural operator learning approach to model biological tissues from digital image correlation measurements. J Biomech Eng 144(12):121012
Estrada JB, Luetkemeyer CM, Scheven UM, Arruda EM (2020) Mr-u: material characterization using 3d displacement-encoded magnetic resonance and the virtual fields method. Exp Mech 60:907–924
Zhang W, Sommer G, Niestrawska JA, Holzapfel GA, Nordsletten D (2022) The effects of viscoelasticity on residual strain in aortic soft tissues. Acta Biomater 140:398–411
Holzapfel GA, Fereidoonnezhad B (2017) Modeling of damage in soft biological tissues. Biomechanics of living organs. Elsevier, Amsterdam, pp 101–123
Vlassis NN, Ma R, Sun W (2020) Geometric deep learning for computational mechanics part i: Anisotropic hyperelasticity. Comput Methods Appl Mech Eng 371:113299
Acknowledgements
VT and IB acknowledge the support of AFOSR under the grant number FA09950-22-1-0061. FSC and MR acknowledge the support of the Open Seed Fund of the School of Engineering at Pontificia Universidad Católica de Chile. ABT acknowledges support from National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institute of Health, United States under award R01AR074525.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no Conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Taç, V., Rausch, M.K., Bilionis, I. et al. Generative hyperelasticity with physics-informed probabilistic diffusion fields. Engineering with Computers 41, 51–69 (2025). https://doi.org/10.1007/s00366-024-01984-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00366-024-01984-2