Abstract
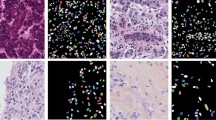
Early detection and the classification of cancer in diagnosed patients can improve the prognosis and improve patient outcomes. In the field of histopathology, the assessment of the disease state is based on the morphological characteristics and spatial distribution of the nuclei in the tissue images. Therefore, the purpose of this research is to propose an automatic histopathological images nuclei segmentation method to accurately predict the boundaries of overlapping and multi-size nuclei. Based on the traditional U-Net, we proposed a convolutional neural network (CNN) that includes iterative attention feature fusion (iAFF) and residual modules for overlapping and multi-size nuclei segmentation task, which is essential and challenging for the development of computer-aided diagnosis (CAD) systems. We extensively evaluate this method on the TNBC and TCGA datasets and the experimental results show that our method can obtain better segmentation performance than the most advanced deep learning models. The proposed method has three advantages in the task of nuclei segmentation. First of all, the iAFF module used in the skip connection fully combines the global channel and the local context and overcomes the semantic and scale inconsistency between the input features. Second, the residual module in the decoder further integrates context information. Third, the method proposed in this paper will not increase too much computational overhead on U-Net, but the effect is significantly improved. Therefore, compared with traditional CNN, multi-level feature fusion network (MFFNet) can reduce redundancy and effectively improve the performance of the model without greatly increasing the network parameters.










Similar content being viewed by others
Availability of data and material
All experimental data in this study are included in this article.
References
Aatresh, A.A., Yatgiri, R.P., Chanchal, A.K., Kumar, A., Ravi, A., Das, D., Raghavendra, B., Lal, S., Kini, J.: Efficient deep learning architecture with dimension-wise pyramid pooling for nuclei segmentation of histopathology images. Comput. Med. Imaging Graph. 93, 101975 (2021)
Chanchal, A.K., Kumar, A., Lal, S., Kini, J.: Efficient and robust deep learning architecture for segmentation of kidney and breast histopathology images. Comput. Electr. Eng. 92, 107177 (2021)
Chen, H., Qi, X., Yu, L., Heng, P.A.: DCAN: deep contour-aware networks for accurate gland segmentation. In: Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, pp. 2487–2496 (2016)
Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder–decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 801–818 (2018)
Cheng, Z., Qu, A., He, X.: Contour-aware semantic segmentation network with spatial attention mechanism for medical image. Vis. Comput. 1–14 (2021)
Chidester, B., Ton, T.V., Tran, M.T., Ma, J., Do, M.N.: Enhanced rotation-equivariant u-net for nuclear segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (2019)
Cruz-Roa, A., Gilmore, H., Basavanhally, A., Feldman, M., Ganesan, S., Shih, N.N., Tomaszewski, J., González, F.A., Madabhushi, A.: Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci. Rep. 7(1), 1–14 (2017)
Dai, Y., Gieseke, F., Oehmcke, S., Wu, Y., Barnard, K.: Attentional feature fusion. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 3560–3569 (2021)
Ding, F., Yang, G., Wu, J., Ding, D., Xv, J., Cheng, G., Li, X.: High-order attention networks for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 253–262. Springer, Berlin (2020)
Fang, W., Han, X.H.: Spatial and channel attention modulated network for medical image segmentation. In: Proceedings of the Asian Conference on Computer Vision (2020)
Hayakawa, T., Prasath, V.S., Kawanaka, H., Aronow, B.J., Tsuruoka, S.: Computational nuclei segmentation methods in digital pathology: a survey. Arch. Comput. Methods Eng. 1–13 (2019)
Jie, S., Liang, X., Mohsen, M., Zhichao, L.: Multi-layer boosting sparse convolutional model for generalized nuclear segmentation from histopathology images. Knowl. Based Syst. 176, 40–53 (2019)
Kang, Q., Lao, Q., Fevens, T.: Nuclei segmentation in histopathological images using two-stage learning. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 703–711. Springer, Berlin (2019)
Kumar, N., Verma, R., Sharma, S., Bhargava, S., Vahadane, A., Sethi, A.: A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 36(7), 1550–1560 (2017)
Li, X., Wang, Y., Tang, Q., Fan, Z., Yu, J.: Dual U-net for the segmentation of overlapping glioma nuclei. IEEE Access 7, 84040–84052 (2019)
Liu, D., Zhang, D., Song, Y., Zhang, C., Zhang, F., O’Donnell. L., Cai, W.: Nuclei segmentation via a deep panoptic model with semantic feature fusion. In: IJCAI, pp 861–868 (2019)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Mahmood, F., Borders, D., Chen, R.J., McKay, G.N., Salimian, K.J., Baras, A., Durr, N.J.: Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 39(11), 3257–3267 (2019)
Naylor, P., Laé, M., Reyal, F., Walter, T.: Nuclei segmentation in histopathology images using deep neural networks. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), pp 933–936. IEEE (2017)
Naylor, P., Laé, M., Reyal, F., Walter, T.: Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans. Med. Imaging 38(2), 448–459 (2018)
Oda, H., Roth, H.R., Chiba, K., Sokolić, J., Mori, K.: Besnet: Boundary-enhanced segmentation of cells in histopathological images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (2018)
Piórkowski, A., Gertych, A.: Color normalization approach to adjust nuclei segmentation in images of hematoxylin and eosin stained tissue. In: International Conference on Information Technologies in Biomedicine, pp. 393–406. Springer, Berlin (2018)
Reinhard, E., Adhikhmin, M., Gooch, B., Shirley, P.: Color transfer between images. IEEE Comput. Graphics Appl. 21(5), 34–41 (2001)
Rojo, M.G.: State of the art and trends for digital pathology. Stud. Health Technol. Inform. 179(179), 15–28 (2012)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241. Springer, Berlin (2015)
Roy, S., Das, D., Lal, S., Kini, J.: Novel edge detection method for nuclei segmentation of liver cancer histopathology images. J. Ambient. Intell. Humaniz. Comput. 1–18,(2021)
Shu, J., Fu, H., Qiu, G., Kaye, P., Ilyas, M.: Segmenting overlapping cell nuclei in digital histopathology images. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp 5445–5448. IEEE (2013)
Sinha, A., Dolz, J.: Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. (2020)
Song, J., Xiao, L., Lian, Z.: Boundary-to-marker evidence-controlled segmentation and mdl-based contour inference for overlapping nuclei. IEEE J. Biomed. Health Inform. 21(2), 451–464 (2015)
Vahadane, A., Peng, T., Sethi, A., Albarqouni, S., Wang, L., Baust, M., Steiger, K., Schlitter, A.M., Esposito, I., Navab, N.: Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 35(8), 1962–1971 (2016)
Veta, M., Van Diest, P.J., Kornegoor, R., Huisman, A., Viergever, M.A., Pluim, J.P.: Automatic nuclei segmentation in h&e stained breast cancer histopathology images. PLoS ONE 8(7), e70221 (2013)
Vuola, A.O., Akram, S.U., Kannala, J.: Mask-RCNN and U-net ensembled for nuclei segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 208–212. IEEE (2019)
Wan, T., Zhao, L., Feng, H., Li, D., Tong, C., Qin, Z.: Robust nuclei segmentation in histopathology using ASPPU-net and boundary refinement. Neurocomputing 408, 144–156 (2020)
Win, K.Y., Choomchuay, S.: Automated segmentation of cell nuclei in cytology pleural fluid images using otsu thresholding. In: 2017 International Conference on Digital Arts, pp. 14–18. Media and Technology (ICDAMT), IEEE (2017)
Yang, L., Ghosh, R.P., Franklin, J.M., Chen, S., You, C., Narayan. R.R., Melcher, M.L., Liphardt, J.T.: Nuset: a deep learning tool for reliably separating and analyzing crowded cells. PLOS Comput. Biol. 16 (2020)
Zeng, Z., Xie, W., Zhang, Y., Lu, Y.: RIC-Unet: An improved neural network based on Unet for nuclei segmentation in histology images. IEEE Access 7, 21420–21428 (2019)
Zhao, J., Dai, L., Zhang, M., Yu, F., Li, M., Li, H., Wang, W., Zhang, L.: Pgu-net+: progressive growing of u-net+ for automated cervical nuclei segmentation. In: International Workshop on Multiscale Multimodal Medical Imaging, pp. 51–58. Springer, Berlin (2019)
Zhao, M., Wang, H., Han, Y., Wang, X., Dai, H.N., Sun, X., Zhang, J., Pedersen, M.: Seens: Nuclei segmentation in pap smear images with selective edge enhancement. Futur. Gener. Comput. Syst. 114, 185–194 (2021)
Acknowledgements
This work is jointly supported by the Natural Science Foundation of Gansu Province (No.18JR3RA288) and the Fundamental Research Funds for the Central Universities of China (No.lzujbky-2017-it72 and No.lzujbky-2018-it61).
Funding
This work is jointly supported by the Natural Science Foundation of Gansu Province (No.18JR3RA288) and the Fundamental Research Funds for the Central Universities of China (No.lzujbky-2017-it72 and No.lzujbky-2018-it61).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Ethical approval
This article does not contain any study with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, X., Pi, J., Lou, M. et al. Multi-level feature fusion network for nuclei segmentation in digital histopathological images. Vis Comput 39, 1307–1322 (2023). https://doi.org/10.1007/s00371-022-02407-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-022-02407-3