Abstract
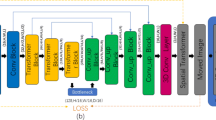
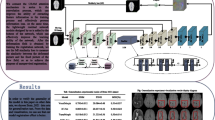
Medical image registration is an essential task in researching and applying medical images. Doctors can observe and extract relevant pathological features to quickly analyze the disease by registered images to diagnose the infection. After more than ten years of research and development, medical image registration has achieved good research results in traditional and deep learning methods. However, most existing methods only focus on unidirectional medical image registration research and rarely consider bidirectional medical image registration research. This paper proposes a new, unsupervised bidirectional medical image registration method based on this aspect. This method guarantees the registration effect in the forward and reverses directions and adds a cascade connection-based channel attention network to the registration model to enable better automatic learning of the registration model, optimizes feature weights, and extracts essential information from images to improve registration performance. We verified the effectiveness of our method by conducting experiments on large-scale 3D brain MRI images and achieved a comparable registration speed and effect with most existing medical image registration methods.











Similar content being viewed by others
References
Sparks, B., Friedman, S., Shaw, D., Aylward, E.H., Echelard, D., Artru, A., Maravilla, K., Giedd, J., Munson, J., Dawson, G.: others: brain structural abnormalities in young children with autism spectrum disorder. Neurology 59, 184–192 (2002)
Andersen, D., Popescu, V., Cabrera, M.E., Shanghavi, A., Gomez, G., Marley, S., Mullis, B., Wachs, J.: Virtual annotations of the surgical field through an augmented reality transparent display. Vis. Comput. 32, 1481–1498 (2016)
Liu, Y., Duan, Y., Zeng, T.: Learning multi-level structural information for small organ segmentation. Signal Process. 193, 108418 (2022)
Zhang, G., Yang, Y., Xu, S., Nan, Y., Lv, C., Wei, L., Qian, T., Han, J., Xie, G.: Autonomous localization and segmentation for body composition quantization on abdominal CT. Biomed. Signal Process. Control 71, 103172 (2022)
Li, Y., Wang, Z., Yin, L., Zhu, Z., Qi, G., Liu, Y.: X-Net: a dual encoding–decoding method in medical image segmentation. Vis. Comput. (2021). https://doi.org/10.1007/s00371-021-02328-7
Jia, F., Wong, W.H., Zeng, T.: DDUNet: Dense dense U-net with applications in image denoising. In: Proceedings of the IEEE/CVF international conference on computer vision. pp. 354–364 (2021)
Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C.: Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008)
Gan, R., Chung, A.C., Liao, S.: Maximum distance-gradient for robust image registration. Med. Image Anal. 12, 452–468 (2008)
Soleimani, M., Aghagolzadeh, A., Ezoji, M.: Symmetry-based representation for registration of multimodal images. Med. Biol. Eng. Comput. 60, 1015–1032 (2022)
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38, 1788–1800 (2019)
Jaderberg, M., Simonyan, K., Zisserman, A., et al.: Spatial transformer networks. Adv. Neural Inf. Process. Syst. 28, (2015)
de Vos, B.D., Berendsen, F.F., Viergever, M.A., Sokooti, H., Staring, M., Isgum, I.: A deep learning framework for unsupervised affine and deformable image registration. Med. Image Anal. 52, 128–143 (2019)
Ahmad, S., Khan, M.F.: Multimodal non-rigid image registration based on elastodynamics. Vis. Comput. 34, 21–27 (2018)
Zhou, C., Cha, T., Peng, Y., Li, G.: Transfer learning from an artificial radiograph-landmark dataset for registration of the anatomic skull model to dual fluoroscopic X-ray images. Comput. Biol. Med. 138, 104923 (2021)
Anzid, H., le Goic, G., Bekkari, A., et al.: A new SURF-based algorithm for robust registration of multimodal images data. Vis. Comput. (2022). https://doi.org/10.1007/s00371-022-02435-z
He, Z., He, Y., Cao, W.: Deformable image registration with attention-guided fusion of multi-scale deformation fields. Appl. Intell. (2022). https://doi.org/10.1007/s10489-022-03659-1
Thirion, J.-P.: Image matching as a diffusion process: an analogy with Maxwell’s demons. Med. Image Anal. 2, 243–260 (1998)
Vercauteren, T., Pennec, X., Perchant, A., Ayache, N.: Diffeomorphic demons: Efficient non-parametric image registration. Neuroimage 45, S61–S72 (2009)
Cui, M., Wonka, P., Razdan, A., Hu, J.: A new image registration scheme based on curvature scale space curve matching. Vis. Comput. 23, 607–618 (2007)
Ashburner, J.: A fast diffeomorphic image registration algorithm. Neuroimage 38, 95–113 (2007)
Dalca, A.V., Balakrishnan, G., Guttag, J., Sabuncu, M.R.: Unsupervised learning for fast probabilistic diffeomorphic registration. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 729–738. Springer (2018)
Rueckert, D., Sonoda, L.I., Hayes, C., Hill, D.L., Leach, M.O., Hawkes, D.J.: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18, 712–721 (1999)
Hellier, P., Ashburner, J., Corouge, I., Barillot, C., Friston, K.J.: Inter-subject registration of functional and anatomical data using SPM. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 590–597. Springer (2002)
Glocker, B., Komodakis, N., Tziritas, G., Navab, N., Paragios, N.: Dense image registration through MRFs and efficient linear programming. Med. Image Anal. 12, 731–741 (2008)
Ou, Y., Sotiras, A., Paragios, N., Davatzikos, C.: DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med. Image Anal. 15, 622–639 (2011)
Rueckert, D., Aljabar, P., Heckemann, R.A., Hajnal, J.V., Hammers, A.: Diffeomorphic registration using B-splines. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 702–709. Springer (2006)
Beg, M.F., Miller, M.I., Trouvé, A., Younes, L.: Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vis. 61, 139–157 (2005)
Rohé, M.-M., Datar, M., Heimann, T., Sermesant, M., Pennec, X.: SVF-Net: learning deformable image registration using shape matching. In: International conference on medical image computing and computer-assisted intervention. pp. 266–274. Springer (2017)
Li, H., Fan, Y.: Non-rigid image registration using fully convolutional networks with deep self-supervision. ArXiv Prepr. ArXiv1709.00799. (2017)
Yang, X., Kwitt, R., Styner, M., Niethammer, M.: Quicksilver: fast predictive image registration–a deep learning approach. Neuroimage 158, 378–396 (2017)
Vos, B.D.D., Berendsen, F.F., Viergever, M.A., Staring, M., Igum, I.: End-to-End unsupervised deformable image registration with a convolutional neural network. Springer, Cham, pp. 204–212 (2017). https://doi.org/10.1007/978-3-319-67558-9_24
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: An unsupervised learning model for deformable medical image registration. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 9252–9260 (2018)
Wu, G., Kim, M., Wang, Q., Shen, D.: Hierarchical attribute-guided symmetric diffeomorphic registration for MR brain images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 90–97. Springer (2012)
Reaungamornrat, S., De Silva, T., Uneri, A., Vogt, S., Kleinszig, G., Khanna, A.J., Wolinsky, J.-P., Prince, J.L., Siewerdsen, J.H.: MIND demons: symmetric diffeomorphic deformable registration of MR and CT for image-guided spine surgery. IEEE Trans. Med. Imaging 35, 2413–2424 (2016)
Altantsetseg, E., Khorloo, O., Konno, K.: Rigid registration of noisy point clouds based on higher-dimensional error metrics. Vis. Comput. 34, 1021–1030 (2018)
Marcus, D.S., Wang, T.H., Parker, J., Csernansky, J.G., Morris, J.C., Buckner, R.L.: Open Access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 19(9), 1498–1507 (2007)
Shattuck, D.W., Mirza, M., Adisetiyo, V., Hojatkashani, C., Salamon, G., Narr, K.L., Poldrack, R.A., Bilder, R.M., Toga, A.W.: Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39, 1064–1080 (2008)
Fischl, B.: FreeSurfer. NeuroImage. 62, 774–781 (2012)
Bottou, L.: Large-scale machine learning with stochastic gradient descent. In: Proceedings of COMPSTAT’2010, pp. 177–186. Springer (2010)
Mok, T., Chung, A.: Fast symmetric diffeomorphic image registration with convolutional neural networks. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2020)
Mok, T.C.W., Chung, A.C.S.: Large deformation diffeomorphic image registration with laplacian pyramid networks. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. pp. 211–221. Springer, Cham (2020)
Avants, B.B., Tustison, N.J., Song, G.: A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011)
Lowe, D.G.: Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60, 91–110 (2004)
Acknowledgements
This work was supported in part by the Yunnan Province Research and development of key technologies for clinical medicine of “heart brain treatment” Project under Grant No.202203AC100052, in part by the National Natural Science Foundation of China under Grant 62202416, Grant 62162068, in part by the Yunnan Province Ten Thousand Talents Program and Yunling Scholars Special Project under Grant YNWR-YLXZ-2018-022, in part by the Yunnan Provincial Science and Technology Department-Yunnan University “Double First-Class” Construction Joint Fund Project under Grant No.2019FY003012.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors declare that there is no conflict of interest regarding the publication of the article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, L., Yang, T., Xie, L. et al. Cascade connection-based channel attention network for bidirectional medical image registration. Vis Comput 39, 5527–5545 (2023). https://doi.org/10.1007/s00371-022-02678-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-022-02678-w