Abstract
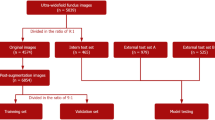
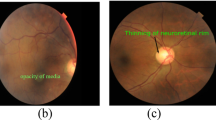
Pancreatic cancer (PC) is an extremely deadly cancer, with mortality rates closely tied to its frequency of occurrence. By the time of diagnosis, pancreatic cancer often presents at an advanced stage, and has often spread to other parts of the body. Due to the poor survival outcomes, PDAC is the fifth leading cause of global cancer death. The 5-year relative survival rate of pancreatic cancer was about 6% and the lowest level in all cancers. Currently, there are no established guidance for screening individuals at high risk for pancreatic cancer, including those with a family history of the pancreatic disease or chronic pancreatitis (CP). With the development of medicine, fundus maps can now predict many systemic diseases. Subsequently, the association between ocular changes and a few pancreatic diseases was also discovered. Therefore, our objective is to construct a deep learning model aimed at identifying correlations between ocular features and significant pancreatic ailments. The utilization of AI and fundus images has extended beyond the investigation of ocular disorders. Hence, in order to solve the tasks of PC and CP classification, we propose a brand new deep learning model (PANet) that integrates pre-trained CNN network, multi-scale feature modules, attention mechanisms, and an FC classifier. PANet adopts a ResNet34 backbone and selectively integrates attention modules to construct its fundamental architecture. To enhance feature extraction capability, PANet combines multi-scale feature modules before the attention module. Our model is trained and evaluated using a dataset comprising 1300 fundus images. The experimental outcomes illustrate the successful realization of our objectives, with the model achieving an accuracy of 91.50% and an area under the receiver operating characteristic curve (AUC) of 96.00% in PC classification, and an accuracy of 95.60% and an AUC of 99.20% in CP classification. Our study establishes a characterizing link between ocular features and major pancreatic diseases, providing a non-invasive, convenient, and complementary method for screening and detection of pancreatic diseases.








Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
No datasets were generated or analyzed during the current study.
References
Al-Jebrni, A.H., Ali, S.G., Li, H., et al.: Sthy-net: a feature fusion-enhanced dense-branched modules network for small thyroid nodule classification from ultrasound images. Vis. Comput. 39(8), 3675–3689 (2023)
Aouaidjia, K., Sheng, B., Li, P., et al.: Efficient body motion quantification and similarity evaluation using 3-D joints skeleton coordinates. IEEE Trans. Syst. Man Cybern. Syst. 51(5), 2774–2788 (2019)
Bahr, T., Vu, T.A., Tuttle, J.J., et al.: Deep learning and machine learning algorithms for retinal image analysis in neurodegenerative disease: systematic review of datasets and models. Transl. Vision Sci. Technol. 13(2), 16–16 (2024)
Campo, S.M.A., Gasparri, V., Catarinelli, G., et al.: Acute pancreatitis with Purtscher’s retinopathy: case report and review of the literature. Dig. Liver Dis. 32(8), 729–732 (2000)
Chari, S.T., Mohan, V., Pitchumoni, C.S., et al.: Risk of pancreatic carcinoma in tropical calcifying pancreatitis: an epidemiologic study. Pancreas 9(1), 62–66 (1994)
Cheema, M.N., Nazir, A., Yang, P., et al.: Modified GAN-cAED to minimize risk of unintentional liver major vessels cutting by controlled segmentation using CTA/SPET-CT. IEEE Trans. Industr. Inf. 17(12), 7991–8002 (2021)
Chen, W., Butler, R.K., Zhou, Y., et al.: Prediction of pancreatic cancer based on imaging features in patients with duct abnormalities. Pancreas 49(3), 413–419 (2020)
Chikumba, S., Hu, Y., Luo, J.: Deep learning-based fundus image analysis for cardiovascular disease: a review. Ther. Adv. Chronic Dis. 14, 20406223231209896 (2023)
Conroy, T., Pfeiffer, P., Vilgrain, V., et al.: Pancreatic cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann. Oncol. 34(11), 987–1002 (2023)
Dai, L., Sheng, B., Chen, T., et al.: A deep learning system for predicting time to progression of diabetic retinopathy. Nat. Med. 30(2), 584–594 (2024)
Dai, L., Wu, L., Li, H., et al.: A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat. Commun. 12(1), 3242 (2021)
Gao, S.H., Cheng, M.M., Zhao, K., et al.: Res2net: a new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 43(2), 652–662 (2019)
Gao, X., Wang, X.: Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: a preliminary study. Diagn. Interv. Imaging 101(2), 91–100 (2020)
Gorris, M., Hoogenboom, S.A., Wallace, M.B., et al.: Artificial intelligence for the management of pancreatic diseases. Dig. Endosc. 33(2), 231–241 (2021)
Guan, Z., Li, H., Liu, R., et al.: Artificial intelligence in diabetes management: advancements, opportunities, and challenges. Cell Rep. Med. 4, 101213 (2023)
He, K., Zhang, X., Ren, S., et al.: Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 770–778 (2016)
Hemelings, R., Elen, B., Schuster, A.K., et al.: A generalizable deep learning regression model for automated glaucoma screening from fundus images. NPJ Digit. Med. 6(1), 112 (2023)
Hu, J.X., Zhao, C.F., Chen, W.B., et al.: Pancreatic cancer: a review of epidemiology, trend, and risk factors. World J. Gastroenterol. 27(27), 4298 (2021)
Huang, G., Liu, Z., Van Der Maaten, L., et al.: Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 4700–4708 (2017)
Huang, S., Liu, X., Tan, T., et al.: TransMRSR: transformer-based self-distilled generative prior for brain MRI super-resolution. Vis. Comput. 39(8), 3647–3659 (2023)
Karambakhsh, A., Sheng, B., Li, P., et al.: SparseVoxNet: 3-D object recognition with sparsely aggregation of 3-D dense blocks. IEEE Trans. Neural Netw. Learn. Syst. 35(1), 532–546 (2022)
Kelly, R.J., Bever, K., Chao, J., et al.: Society for immunotherapy of cancer (SITC) clinical practice guideline on immunotherapy for the treatment of gastrointestinal cancer. J. Immunother. Cancer 11(6), e006658 (2023)
Koonce, B., Koonce, B.E.: Convolutional neural networks with swift for tensorflow: image recognition and dataset categorization. Apress, New York (2021)
Kumar, K.V., Soora, N.R., Santoshkumar, N.C.: Fundus image classification for the early detection of issues in the DR for the effective disease diagnosis. J. Comput. Allied Intell. 1(01), 27–40 (2023)
Lee, H.A., Chen, K.W., Hsu, C.Y.: Prediction model for pancreatic cancer—a population-based study from NHIRD. Cancers 14(4), 882 (2022)
Li, J., Zhang, P., Wang, T., et al.: DSMT-Net: dual self-supervised multi-operator transformation for multi-source endoscopic ultrasound diagnosis. IEEE Trans. Med. Imaging 43, 64–75 (2023)
Li, P., Liang, L., Gao, Z., et al.: AMD-Net: automatic subretinal fluid and hemorrhage segmentation for wet age-related macular degeneration in ocular fundus images. Biomed. Signal Process. Control 80, 104262 (2023)
Li, T., Bo, W., Hu, C., et al.: Applications of deep learning in fundus images: a review. Med. Image Anal. 69, 101971 (2021)
Li, Y., Zhang, R., Dong, L., et al.: Predicting systemic diseases in fundus images: systematic review of setting, reporting, bias, and models’ clinical availability in deep learning studies. Eye 38, 1246–1251 (2024)
Lim, G., Lim, Z.W., Xu, D., et al. Feature isolation for hypothesis testing in retinal imaging: an ischemic stroke prediction case study. In: Proceedings of the AAAI conference on artificial intelligence 33(1): 9510–9515 (2019)
Liu, R., Ou, L., Sheng, B., et al.: Mixed-weight neural bagging for detecting $ m^ 6A $ modifications in SARS-CoV-2 RNA sequencing. IEEE Trans. Biomed. Eng. 69(8), 2557–2568 (2022)
Liu, R., Wang, X., Wu, Q., et al.: DeepDRiD: diabetic retinopathy-grading and image quality estimation challenge. Patterns 3(6), 100512 (2022)
Manikandan, J., Krishna, B.V., Sasivarma, C., et al.: Cataract fundus image detection using hybrid deep learning model. In: International conference on computational intelligence in data science. Cham: Springer Nature Switzerland pp. 300–313 (2023)
Mayer, C., Khoramnia, R.: Purtscher-like retinopathy caused by acute pancreatitis. The Lancet 378(9803), 1653 (2011)
McGuigan, A., Kelly, P., Turkington, R.C., et al.: Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 24(43), 4846 (2018)
Midha, S., Chawla, S., Garg, P.K.: Modifiable and non-modifiable risk factors for pancreatic cancer: a review. Cancer Lett. 381(1), 269–277 (2016)
Mitani, A., Huang, A., Venugopalan, S., et al.: Detection of anaemia from retinal fundus images via deep learning. Nat. Biomed. Eng. 4(1), 18–27 (2020)
Nazir, A., Cheema, M.N., Sheng, B., et al.: Living donor-recipient pair matching for liver transplant via ternary tree representation with cascade incremental learning. IEEE Trans. Biomed. Eng. 68(8), 2540–2551 (2021)
Pandey, R., Rana, S.S., Gupta, V., et al.: Retino-choroidal changes in patients with acute pancreatitis: a prospective analysis of a novel biomarker. Pancreatology 20(8), 1604–1610 (2020)
Paul, W., Burlina, P., Mocharla, R., et al.: Accuracy of artificial intelligence in estimating best-corrected visual acuity from fundus photographs in eyes with diabetic macular edema. JAMA Ophthalmol. 141(7), 677–685 (2023)
Placido, D., Yuan, B., Hjaltelin, J.X., et al.: A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 29(5), 1113–1122 (2023)
Qian, B., Chen, H., Wang, X., et al.: DRAC 2022: a public benchmark for diabetic retinopathy analysis on ultra-wide optical coherence tomography angiography images. Patterns (2024). https://doi.org/10.1016/j.patter.2024.100929
Rahim, S., Sabri, K., Ells, A., et al.: Novel fundus image preprocessing for Retcam images to improve deep learning classification of retinopathy of prematurity. (2023) https://doi.org/10.48550/arXiv.2302.02524
Rim, T.H., Lee, G., Kim, Y., et al.: Prediction of systemic biomarkers from retinal photographs: development and validation of deep-learning algorithms. Lancet Digit. Health 2(10), e526–e536 (2020)
Sabanayagam, C., Xu, D., Ting, D.S.W., et al.: A deep learning algorithm to detect chronic kidney disease from retinal photographs in community-based populations. Lancet Digit. Health 2(6), e295–e302 (2020)
Shen, Z., Savvides, M.: Meal v2: boosting vanilla resnet-50 to 80%+ top-1 accuracy on imagenet without tricks (2020) https://doi.org/10.48550/arXiv.2009.08453
Sheng, B., Guan, Z., Lim, L.L., et al.: Large language models for diabetes care: potentials and prospects. Sci. Bull. S2095–9273(24), 00004 (2024)
Simonyan K, Zisserman, A.: Very deep convolutional networks for large-scale image recognition. (2014) https://doi.org/10.48550/arXiv.1409.1556
Steiner, M., del Mar, E.-O., Muñoz-Fernández, S.: Choroidal and retinal thickness in systemic autoimmune and inflammatory diseases: a review[J]. Surv. Ophthalmol. 64(6), 757–769 (2019)
Szegedy, C., Ioffe, S., Vanhoucke, V., et al.: Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence (2017)
Tan, Y., Ma, Y., Rao, S., et al.: Performance of deep learning for detection of chronic kidney disease from retinal fundus photographs: a systematic review and meta-analysis. Eur. J. Ophthalmol. 34(2), 502–509 (2024)
Ting, D.S.W., Peng, L., Varadarajan, A.V., et al.: Deep learning in ophthalmology: the technical and clinical considerations. Prog. Retin. Eye Res. 72, 100759 (2019)
Viedma, I.A., Alonso-Caneiro, D., Read, S.A., et al.: Deep learning in retinal optical coherence tomography (OCT): a comprehensive survey. Neurocomputing 507, 247–264 (2022)
Visioli, G., Zeppieri, M., Iannucci, V., et al.: From bedside to diagnosis: the role of ocular fundus in systemic infections. J. Clin. Med. 12(23), 7216 (2023)
Wang, Q., Wu, B., Zhu, P., et al.: ECA-Net: efficient channel attention for deep convolutional neural networks. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 11534–11542 (2020)
Wong, T.Y., Klein, R., Couper, D.J., et al.: Retinal microvascular abnormalities and incident stroke: the atherosclerosis risk in communities study. Lancet 358(9288), 1134–1140 (2001)
Xiao, W., Huang, X., Wang, J.H., et al.: Screening and identifying hepatobiliary diseases through deep learning using ocular images: a prospective, multicentre study. Lancet Digit. Health 3(2), e88–e97 (2021)
Xu, P.P., Liu, T.Y., Zhou, F., et al.: Artificial intelligence in coronary computed tomography angiography. Med. Plus 10, 100001 (2023)
Zhu, X., Xiong, Y., Dai, J., et al.: Deep feature flow for video recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 2349–2358 (2017)
Zou, K.H., O’Malley, A.J., Mauri, L.: Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 115(5), 654–657 (2007)
Acknowledgements
The authors would like to thank the participants of the study as well as Shanghai Sixth’s People’s Hospital and Shanghai Jiao Tong University for providing funding and organizational support. Many thanks to Tingting Li and Xuan Cai, two attending ophthalmologist, who have been practicing ophthalmology for more than 10 years, for reading fundus images in this study.
Funding
This work was supported by the College-level Project Fund of Shanghai Sixth People’s Hospital (Grant No. ynlc201909) and the Interdisciplinary Program of Shanghai Jiao Tong University (Project No. YG2022QN089).
Author information
Authors and Affiliations
Contributions
Y. W. and P. F. contributed equally to this work. Corresponding author: J. S. (Email: slyysj2009@163.com). All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Fang, P., Wang, X. et al. Predicting pancreatic diseases from fundus images using deep learning. Vis Comput 41, 3553–3564 (2025). https://doi.org/10.1007/s00371-024-03619-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-024-03619-5