Abstract
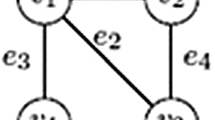
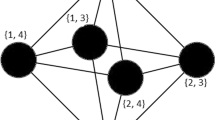
We consider a class of pattern graphs on \(q\ge 4\) vertices that have \(q-2\) distinguished vertices with equal neighborhood in the remaining two vertices. Two pattern graphs in this class are siblings if they differ by some edges connecting the distinguished vertices. In particular, we show that if induced copies of siblings to a pattern graph in such a class are rare in the host graph then one can detect the pattern graph relatively efficiently. For an example, we infer that if there are \(N_d\) induced copies of a diamond (i.e., a graph on four vertices missing a single edge to be complete) in the host graph, then an induced copy of the complete graph on four vertices, \(K_4,\) as well as an induced copy of the cycle on four vertices, \(C_4,\) can be deterministically detected in \(O(n^{2.75}+N_d)\) time. Note that the fastest known algorithm for \(K_4\) and the fastest known deterministic algorithm for \(C_4\) run in \(O(n^{3.257})\) time. By using random bits, we can speed up our method such that the number of induced copies of the siblings is replaced by the ratio of this number to the number of induced copies of the pattern graph plus 1 in the upper time bound. We also show that if there is a family of siblings whose induced copies in the host graph are rare then there are good chances to determine the numbers of occurrences of induced copies for all pattern graphs on q vertices relatively efficiently.


Similar content being viewed by others
Notes
In fact, \(\omega (1,1,2)=\omega (1,2,1)=\omega (2,1,1)\) holds [10].
For simplicity, we shall refer to a subgraph of a graph G that is isomorphic to a graph H as a copy of H in G or just an H copy in G.
References
Alon, N., Dao, P., Hajirasouliha, I., Hormozdiari, F., Sahinalp, S.C.: Biomolecular network motif counting and discovery by color coding. Bioinformatics (ISMB 2008) 24(13), 241–249 (2008)
Bläser, M., Komarath, B., Sreenivasaiah, K.: Graph pattern polynomials. In: Proceedings of 38th IARCS Annual Conference on Foundations of Software Technology and Theoretical Computer Science (FSTTCS), pp. 18:1–18:13 (2018)
Corneil, D.G., Perl, Y., Stewart, L.K.: A linear recognition algorithm for cographs. SIAM J. Comput. 14(4), 926–934 (1985)
Dalirrooyfard, M., Duong Vuong, T., Virginia Vassilevska Williams, V.: Graph pattern detection: hardness for all induced patterns and faster non-induced cycles. In: Proceedings of 51st Annual ACM Symposium on Theory of Computing (STOC), pp. 1167–1178 (2019)
Eisenbrand, F., Grandoni, F.: On the complexity of fixed parameter clique and dominating set. Theoret. Comput. Sci. 326, 57–67 (2004)
Eschen, E.M., Hoàng, C.T., Spinrad, J., Sritharan, R.: On graphs without a C4 or a diamond. Discrete Appl. Math. 159(7), 581–587 (2011)
Floderus, P., Kowaluk, M., Lingas, A., Lundell, E.-M.: Detecting and counting small pattern graphs. SIAM J. Discrete Math. 29(3), 1322–1339 (2015)
Floderus, P., Kowaluk, M., Lingas, A., Lundell, E.-M.: Induced subgraph isomorphism: are some patterns substantially easier than others? Theoret. Comput. Sci. 605, 119–128 (2015)
Gąsieniec, L., Kowaluk, M., Lingas, A.: Faster multi-witnesses for Boolean matrix product. Inf. Process. Lett. 109, 242–247 (2009)
Huang, X., Pan, V.Y.: Fast rectangular matrix multiplications and applications. J. Complex. 14, 257–299 (1998)
Itai, A., Rodeh, M.: Finding a minimum circuit in a graph. SIAM J. Comput. 7, 413–423 (1978)
Kloks, T., Kratsch, D., Müller, H.: Finding and counting small induced subgraphs efficiently. Inf. Process. Lett. 74(3–4), 115–121 (2000)
Kowaluk, M., Lingas, A., Lundell, E.-M.: Counting and detecting small subgraphs via equations. SIAM J. Discrete Math. 27(2), 892–909 (2013)
Kowaluk, M., Lingas, A.: A Fast deterministic detection of small pattern graphs in graphs without large cliques. In: Proceedings of the 11th International Conference and Workshops (WALCOM 2017), LNCS 10167, pp. 217–227. Springer, Switzerland (2017)
Le Gall, F.: Faster algorithms for rectangular matrix multiplication. In: Proceedings of 53rd Symposium on Foundations of Computer Science (FOCS), pp. 514–523 (2012)
Le Gall, F.: Powers of tensors and fast matrix multiplication. In: Proceedings of 39th International Symposium on Symbolic and Algebraic Computation, pp. 296–303 (2014)
Nes̆etr̆il, J., Poljak, S.: On the complexity of the subgraph problem. Comment. Math. Univ. Carol. 26(2), 415–419 (1985)
Olariu, S.: Paw-free graphs. Inf. Process. Lett. 28, 53–54 (1988)
Schank, T., Wagner, D.: Finding, counting and listing all triangles in large graphs, an experimental study. In: Proceedings of WEA, pp. 606–609 (2005)
Sekar, V., Xie, Y., Maltz, D.A., Reiter, M.K, Zhang, H.: Toward a framework for internet forensic analysis. In: Third Workshop on Hot Topics in Networking (HotNets-HI) (2004)
Wolinski, C., Kuchcinski, K., Raffin, E.: Automatic design of application-specific reconfigurable processor extensions with UPaK synthesis kernel. ACM Trans. Des. Autom. Electron. Syst. 15(1), 1–36 (2009)
Williams, V.V., Wang, J.R., Williams, R., Yu H.: Finding four-node subgraphs in triangle Time. In: Proceedings of SODA, pp. 1671–1680 (2015)
Williams, V.V.: Multiplying matrices faster than Coppersmith-Winograd. In: Proceedings of 44th Annual ACM Symposium on Theory of Computing (STOC), pp. 887–898 (2012)
Acknowledgements
The authors are thankful to anonymous referees for valuable comments. The research has been supported in part by Swedish Research Council grant 621-2017-03750.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A preliminary, short version of this paper has appeared in Proceedings of 22nd Symposium on Fundamentals of Computation Theory (FCT 2019), Lecture Notes in Computer Science 11651, Copenhagen, Denmark, August 12–14, 2019.
Appendix: Proof of Lemma 2.6
Appendix: Proof of Lemma 2.6
1.1 A Key Fact
Fact 5.1
(Theorem 4.1 in [13]) If for all \(H\in H_k\setminus H_k(l)\) the values NI(H, G) are known then for all \(H''\in H_k,\) the numbers \(NI(H'',G)\) can be determined in time \(O(|H_k(l)|(n^{l}(k-l)+|H_k|k^2k!+|H_k(l)|^2)+T_l(n)),\) in particular in time \(O(n^{l}+T_l(n))\) for \(k=O(1).\) (Recall that \(T_l(n)\) stands for the time required to solve the l-neighborhood problem.)
1.2 Proof of the Lemma
Consider Fact 5.1 (Theorem 4.1 in [13]) with fixed \(k=q\), \(l=q-2\), and \(O(n^{\omega (\lceil (q-2)/2 \rceil ,1,\lfloor (q-2)/2 \rfloor })\) substituted for \(T_{q-2}(n)\) according to (already described) Theorem 6.1 in [13]. It follows that if for all \(H\in \mathcal {H}_{q-2}\setminus \mathcal {H}_{q-2}(q-2)\) the values NI(H, G) are known then for all \(H'\in \mathcal {H}_{q-2},\) the numbers \(NI(H',G)\) can be determined in time \(O(n^{\omega (\lceil (q-2)/2 \rceil ,1,\lfloor (q-2)/2 \rfloor })\). Since \(H_q\setminus H_q(q-2)=\{K_q\}\), it follows directly from Fact 5.1 that if the number of (induced) subgraphs isomorphic to \(K_q\) in the host graph is known then for all the pattern graphs on q vertices the corresponding numbers can be computed in the time specified in the lemma statement. The argumentation given in the proof of Fact 5.1 (Theorem 4.1 in [13]) works equally well when the number of induced copies of an arbitrary pattern graph H on q vertices is known.
Namely, following the proof of Theorem 4.1 in [13] with \(k=q\) and \(l=q-2,\) we form \(SEq(G,q,q-2).\) Since we assume that q is fixed, the coefficients \(B(H_{sub},H')\) on the left-sides of the equations in \(SEq(G,q,q-2)\) can be computed in O(1) time. By (already described) Lemma 3.5 and Theorem 6.1 in [13], the right-sides of the equations can be computed in time \(O(n^{\omega (\lceil (q-2)/2 \rceil ,1,\lfloor (q-2)/2 \rfloor }) .\) Let us order the graphs in \(\mathcal {H}_k\) so that the number of edges is non-decreasing and the graphs in \(\mathcal {H}_k(l)\) form a prefix of the sorted sequence. Let B be the \(|\mathcal {H}_k(l)|\times |\mathcal {H}_k|\) matrix corresponding to the left-hand sides of the equations in \(SEq(G,q,q-2)\), with the rows of B corresponding to \(H\in \mathcal {H}_k(q-2)\) and the columns of B corresponding to \(H'\in \mathcal {H}_{q}\) sorted in the aforementioned way. Consider the leftmost maximal square submatrix M of the matrix B. Since M has zeros below the diagonal starting from the leftmost top-left corner, we infer that the resulting \(|H_q(q-2)|\) equations with \(|H_q|\) unknowns are also linearly independent. Hence, when we substitute the known number of induced copies of H for the variable \(x_{H,G}\) corresponding to H in the equations, we obtain a system S of \(|H_q(q-2)|\) equations with \(|H_q|-1\) unknowns. Since \(H_q\setminus H_q(q-2)=\{K_q\}\), the number of unknowns is equal to the number of equations. It follows from Remark 2.5 that the system S of equations resulting from the substitution is independent. Also, none of the resulting equations can disappear after the substitution. Hence, we can solve them in \(O(|H_q(q-2)|^3)=O(1)\) time. \(\square \)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kowaluk, M., Lingas, A. Rare Siblings Speed-Up Deterministic Detection and Counting of Small Pattern Graphs. Algorithmica 85, 976–991 (2023). https://doi.org/10.1007/s00453-022-01063-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00453-022-01063-2