Abstract
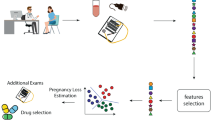
Ovarian cancer is one of the most common types of cancer in women. Correct differentiation between benign and malignant ovarian tumors is of immense importance in medical fields. In this paper, we introduce group penalized logistic regressions to enhance diagnosis accuracy. Firstly, we divide 349 ovarian cancer patients into two sets: one for learning model parameters, and the other for assessing prediction performance, and select 46 variables from 49 traits as the predictor vector to construct GLASSO/GSCAD/GMCP penalized logistic regressions with 11 groups. Secondly, we develop group coordinate descent (GCD) algorithm and its specific pseudo code to simultaneously complete group selection and group estimation, introduce the tenfold cross validation (CV) procedure to select the relatively optimal tuning parameter, and apply the testing set and Youden index to obtain class probability estimator and class index information. Finally, we compute the accuracy, precision, specificity, sensitivity, F1-score and the area under ROC curve (AUC) to assess the prediction performance to the proposed group penalized methods, and found that GLASSO/GSCAD/GMCP penalized logistic regressions outperform three machine learning methods (ANN, artificial neural network; SVM, support vector machine; XGBoost, eXtreme gradient boosting) and three deep learning methods (CNN, convolutional neural network; DNN, deep neural network; RNN, recurrent neural network) in terms of accuracy, F1-score and AUC.




Similar content being viewed by others
Data availability
The data sets used in this study are available from https://www.kaggle.com/saurabhshahane/predict-ovarian-cancer.
References
Alam TM, Shaukat K, Khan WA, Hameed IA, Almuqren LA, Raza MA, Aslam M, Luo S (2022) An efficient deep learning-based skin cancer classifier for an imbalanced dataset. Diagnostics (Basel) 12(9):2115. https://doi.org/10.3390/diagnostics12092115
Alwakid G, Gouda W, Humayun M, Sama NU (2022) Melanoma detection using deep learning-based classifications. Healthcare (Basel) 10(12):2481. https://doi.org/10.3390/healthcare10122481
Anton C, Carvalho FM, Oliveira EI, Maciel GAR, Baracat EC, Carvalho JP (2012) A comparison of CA125, HE4, risk ovarian malignancy algorithm (ROMA), and risk malignancy index (RMI) for the classification of ovarian masses. Clinics (Sao Paulo) 67(5):437–441. https://doi.org/10.6061/clinics/2012(05)06
Bassel A, Abdulkareem AB, Alyasseri ZAA, Sani NS, Mohammed HJ (2022) Automatic malignant and benign skin cancer classification using a hybrid deep learning approach. Diagnostics (Basel) 12(10):2472. https://doi.org/10.3390/diagnostics12102472
Breheny P, Huang J (2011) Coordinate descent algorithms for nonconvex penalized regression, with applications to biological feature selection. Ann Appl Stat 5(1):232–253. https://doi.org/10.1214/10-AOAS388
Breheny P, Huang J (2015) Group descent algorithms for nonconvex penalized linear and logistic regression models with grouped predictors. Stat Comput 25(2):173–187. https://doi.org/10.1007/s11222-013-9424-2
Chen H, Xiang Y (2017) The study of credit scoring model based on group LASSO. Procedia Comput Sci 122:677–684. https://doi.org/10.1016/j.procs.2017.11423
Chen W, Jiang MR, Zhang WG, Chen ZS (2021) A novel graph convolutional feature based convolutional neural network for stock trend prediction. Inf Sci 556:67–94. https://doi.org/10.1016/j.ins.2020.12.068
D’Angelo G, Palmieri F (2023) A co-evolutionary genetic algorithm for robust and balanced controller placement in software-defined networks. J Netw Comput Appl 212:103583. https://doi.org/10.1016/j.jnca.2023.103583
D’Angelo G, Scoppettuolo MN, Cammarota AL, Rosati A, Palmieri F (2022) A genetic programming-based approach for classifying pancreatic adenocarcinoma: the SICED experience. Soft Comput 26:10063–10074. https://doi.org/10.1007/s00500-022-07383-3
D’Angelo G, Della-Morte D, Pastore D, Donadel G, Stefano AD, Palmieri F (2023) Identifying patterns in multiple biomarkers to diagnose diabetic foot using an explainable genetic programming-based approach. Futur Gener Comput Syst 140:138–150. https://doi.org/10.1016/j.future.2022.10.019
Demšar J (2006) Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 7:1–30
Díaz-Padilla I, Razak ARA, Minig L, Bernardini MQ, del Campo JM (2012) Prognostic and predictive value of CA-125 in the primary treatment of epithelial ovarian cancer: potentials and pitfalls. Clin Transl Oncol 14(1):15–20. https://doi.org/10.1007/s12094-012-0756-8
Fan J, Li R (2001) Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc 96(456):1348–1360. https://doi.org/10.1198/016214501753382273
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22. https://doi.org/10.18637/jss.v033.i01
Ghosal R, Maity A, Clark T, Longo SB (2020) Variable selection in functional linear concurrent regression. Appl Stat 69(3):565–587. https://doi.org/10.1111/rssc.12408
Huang J, Breheny P, Ma S (2012) A selective review of group selection in high-dimensional models. Stat Sci Rev J Inst Math Stat 27(4):481–499. https://doi.org/10.1214/12-STS392
Kikkawa F, Nawa A, Tamakoshi K, Ishikawa H, Kuzuya K, Suganuma N, Hattori S, Furui K, Kawai M, Arii Y (1998) Diagnosis of squamous cell carcinoma arising from mature cystic teratoma of the ovary. Cancer 82(11):2249–2255. https://doi.org/10.1002/(SICI)1097-0142(19980601)82:11%3c2249::AID-CNCR21%3e3.0.CO;2-T
Liu X, Goncalves AR, Cao P, Zhao D, Banerjee A (2018) Modeling Alzheimer’s disease cognitive scores using multi-task sparse group LASSO. Comput Med Imaging Graph 66:100–114. https://doi.org/10.1016/j.compmedimag.2017.11.001
Long J, Chen Z, He W, Wu T, Ren J (2020) An integrated framework of deep learning and knowledge graph for prediction of stock price trend: an application in Chinese stock exchange market. Appl Soft Comput 91(4):106205. https://doi.org/10.1016/j.asoc.2020.106205
Lu M, Fan Z, Xu B, Chen L, Zheng X, Li J, Znati T, Mi Q, Jiang J (2020) Using machine learning to predict ovarian cancer. Int J Med Inf. https://doi.org/10.1016/j.ijmedinf.2020.104195
Mazumder R, Friedman JH, Hastie T (2011) Sparsenet: coordinate descent with nonconvex penalties. J Am Stat Assoc 106(495):1125–1138. https://doi.org/10.1198/jasa.2011.tm09738
Meier L, van de Geer S, Bühlmann P (2008) The group lasso for logistic regression. J Roy Stat Soc B 70(1):53–71. https://doi.org/10.1111/j.1467-9868.2007.00627.x
Muinao T, Boruah HPD, Pal M (2019) Multi-biomarker panel signature as the key to diagnosis of ovarian cancer. Heliyon. https://doi.org/10.1016/j.heliyon.2019.e02826
Raghavan R, Ashour FS, Bailey R (2016) A review of cutoffs for nutritional biomarkers. Adv Nutr 7(1):112–120. https://doi.org/10.3945/an.115.009951
Robbins CL, Whiteman MK, Hillis SD, Curtis KM, McDonald JA, Wingo PA, Kulkarni A, Marchbanks PA (2009) Influence of reproductive factors on mortality after epithelial ovarian cancer diagnosis. Cancer Epidemiol Biomark Prev 18(7):2035–2041. https://doi.org/10.1158/1055-9965.EPI-09-0156
Shimizu Y, Yoshimoto J, Toki S, Takamura M, Yoshimura S, Okamoto Y, Yamawaki S, Doya K (2015) Toward probabilistic diagnosis and understanding of depression based on functional MRI data analysis with logistic group LASSO. PLoS ONE. https://doi.org/10.1371/journal.pone.0123524
Tibshirani R (1996) Regression shrinkage and selection via the LASSO. J Roy Stat Soc B 58(1):267–288. https://doi.org/10.1111/j.1467-9868.2011.00771.x
Vincent M, Hansen NR (2014) Sparse group LASSO and high dimensional multinomial classification. Comput Stat Data Anal 71:771–786. https://doi.org/10.48550/arXiv.1205.1245
Wang J, Gao J, Yao H, Wu Z, Wang M, Qi J (2014) Diagnostic accuracy of serum HE4, CA125 and ROMA in patients with ovarian cancer: a meta-analysis. Tumor Biol 35(6):6127–6138. https://doi.org/10.1007/s13277-014-1811-6
Wei FR, Zhu HX (2012) Group coordinate descent algorithms for nonconvex penalized regression. Comput Stat Data Anal 56(2):316–326. https://doi.org/10.1016/j.csda.2011.08.007
Yuan M, Lin Y (2006) Model selection and estimation in regression with grouped variables. J Roy Stat Soc Ser B (stat Methodol) 68(1):49–67. https://doi.org/10.1111/j.1467-9868.2005.00532.x
Zhang C (2010) Nearly unbiased variable selection under minimax concave penalty. Ann Stat 38(2):894–942. https://doi.org/10.2307/25662264
Zhao J, Zeng D, Liang S, Kang H, Liu Q (2021) Prediction model for stock price trend based on recurrent neural network. J Ambient Intell Humaniz Comput 12:745–753. https://doi.org/10.1007/s12652-020-02057-0
Zhu Q, Mao Z, Chen G (2023) Analysis of relationship between tumor markers and detection of tumors by chemiluminescence immunoassay and artificial neural networks. Soft Comput. https://doi.org/10.1007/s00500-023-08855-w
Funding
Hu’s research was supported by the Fifth Batch of Excellent Talent Support Program of Chongqing Colleges and University (68021900601), the Natural Science Foundation of CQ CSTC (cstc.2018jcyjA2073), the Program for the Chongqing Statistics Postgraduate Supervisor Team (yds183002), Chongqing Social Science Plan Project (2019WT59), Science and Technology Research Program of Chongqing Education Commission (KJZD-M202100801), Mathematic and Statistics Team from Chongqing Technology and Business University (ZDPTTD201906) and Open Project from Chongqing Key Laboratory of Social Economy and Applied Statistics (KFJJ2022056).
Author information
Authors and Affiliations
Contributions
XH provided the basic idea, improved the initial writing, and completed the main revisions. YX collected data, provided the figures and the tables, and completed the initial writing and the part revisions. YY and HJ took part in the program writing for the original version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethic approval
This is an observational study and does not require ethics approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, X., Xie, Y., Yang, Y. et al. Group penalized logistic regression differentiates between benign and malignant ovarian tumors. Soft Comput 27, 18565–18584 (2023). https://doi.org/10.1007/s00500-023-09231-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-023-09231-4