Abstract
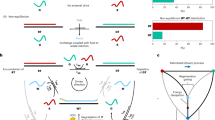
The uncertain and inexact nature of the chemical reactions used to implement DNA computations can be turned into an advantage for implementing robust soft computing systems. The key feature of DNA hybridization that makes it appropriate for fuzzy computing is the uncertainty and incompleteness in the formation of a double-stranded duplex from single-stranded oligonucleotides. To implement fuzzy computing, a set of encoding DNA molecules is given that reproduces a specific membership function in the energetics of the DNA duplex. In addition, a fuzzy inference system implemented with DNA hybridization on solid supports is discussed. The ultimate success of this idea as a general technique, however, is dependent on the actual geometry of the Gibbs free-energy landscapes in the space of all duplex formations. Elucidating this problem is undoubtedly of great importance for biomolecular implementation of soft-computing because it may, in particular, shed light on the true import of fuzzy models in biological processes fundamental to life.
Similar content being viewed by others
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Deaton, R., Garzon, M. Fuzzy logic with biomolecules. Soft Computing 5, 2–9 (2001). https://doi.org/10.1007/s005000000060
Issue Date:
DOI: https://doi.org/10.1007/s005000000060