Abstract
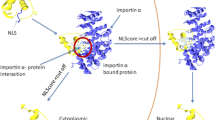
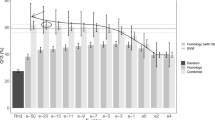
Nuclear export signal (NES) is a nuclear targeting signal within cargo proteins, which is involved in signal transduction and cell cycle regulation. NES is believed to be “born to be weak”; hence, it is a challenge in computational biology to identify it from high-throughput data of amino acid sequences. This work endeavors to tackle the challenge by proposing a computational approach to identifying NES using spiking neural P (SN P) systems. Specifically, secondary structure elements of 30 experimentally verified NES are randomly selected for training an SN P system, and then 1224 amino acid sequences (containing 1015 regular amino acid sequences and 209 experimentally verified NES) abstracted from 221 NES-containing protein sequences randomly in NESdb are selected to test our method. Experimental results show that our method achieves a precision rate 75.41 %, better than NES-REBS 47.2 %, Wregex 25.4 %, ELM, and NetNES 37.4 %. The results of this study are promising in terms of the fact that it is the first feasible attempt to use SN P systems in computational biology after many theoretical advancements.





Similar content being viewed by others
References
Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15:607–660
Conti E, Izaurralde E (2001) Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol 13:310–319
Ren X-X, Wang H-B, Li C, Jiang J-F, Xiong S-D, Jin X, Wu L, Wang J-H (2016) HIV-1 nef-associated factor 1 enhances viral production by interacting with CRM1 to promote nuclear export of unspliced HIV-1 gag mRNA. J Biol Chem 291:4580–4588
Weis K (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441–451
Strambio-De-Castillia C, Niepel M, Rout MP (2010) The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11:490–501
La Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536
Fischer U, Huber J, Boelens WC, Mattajt LW, Lührmann R (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475–483
Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G (1994) Evidence that HIV-1 rev directly promotes the nuclear export of unspliced RNA. EMBO J 13:4105
Ho JH-N, Kallstrom G, Johnson AW (2000) Nmd3p is a crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151:1057–1066
Vissinga CS, Yeo TC, Warren S, Brawley JV, Phillips J, Cerosaletti K, Concannon P (2009) Nuclear export of NBN is required for normal cellular responses to radiation. Mol Cell Biol 29:1000–1006
Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997) Crm1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060
KIrlI K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, Urlaub H, Görlich D (2016) A deep proteomics perspective on crm1-mediated nuclear export and nucleocytoplasmic partitioning. eLife e11466. doi:10.7554/eLife.11466
Xu D, Grishin NV, Chook YM (2012) NESdb: a database of nes-containing crm1 cargoes. Mol Biol Cell 23:3673
Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Travé G, Gibson TJ (2008) Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 13:6580–6603
Iraia G-S, Sonia B, Jose AR (2012) A global survey of crm1-dependent nuclear export sequences in the human deubiquitinase family. Biochem J 441:209–217
Via A, Gould CM, Gemünd C, Gibson TJ, Helmer-Citterich M (2009) A structure filter for the eukaryotic linear motif resource. BMC Bioinform 10:351
Lee T-Y, Lin Z-Q, Hsieh S-J, Bretaña NA, Lu C-T (2011) Exploiting maximal dependence decomposition to identify conserved motifs from a group of aligned signal sequences. Bioinformatics 27:1780–1787
Van Berlo RJ, Wessels LF, De Ridder D, Reinders MJ (2007) Protein complex prediction using an integrative bioinformatics approach. J Bioinform Comput Biol 5:839–864
la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S (2003) NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res 31:393–396
Xu D, Farmer A, Collett G, Grishin NV, Chook YM (2012) Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell 23:3677–3693
Dong X, Biswas A, Chook YM (2009) Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol 16:558–560
Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D (2010) NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol 17:1367–1376
Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C et al (2009) ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. doi:10.1093/nar/gkp1016
Fu S-C, Imai K, Horton P (2011) Prediction of leucine-rich nuclear export signal containing proteins with nessential. Nucleic Acids Res. doi:10.1093/nar/gkr493
Prieto G, Fullaondo A, Rodriguez JA (2014) Prediction of nuclear export signals using weighted regular expressions (wregex). Bioinformatics 30(9):1220–1227. doi:10.1093/bioinformatics/btu016
Ionescu M, Păun G, Yokomori T (2006) Spiking neural P systems. Fundam Inform 71:279–308
Maass W (1997) Networks of spiking neurons: the third generation of neural network models. Neural Networks 10:1659–1671
Chen H, Freund R, Ionescu M, Păun G, Pérez-Jiménez MJ (2007) On string languages generated by spiking neural P systems. Fundam Inform 75:141–162
Păun A, Păun G (2007) Small universal spiking neural P systems. BioSyst 90:48–60
Pan L, Paun G (2009) Spiking neural P systems with anti-spikes. Int J Comput Commun Control IV(3):273–282
Song T, Pan L, Jiang K, Song B, Chen W (2013) Normal forms for some classes of sequential spiking neural P systems. IEEE Trans NanoBiosci 12:255–264
Cavaliere M, Ibarra OH, Păun G, Egecioglu O, Ionescu M, Woodworth S (2009) Asynchronous spiking neural P systems. Theor Comput Sci 410:2352–2364
Song T, Pan L, Păun G (2012) Asynchronous spiking neural P systems with local synchronization. Inf Sci 219:197–207
Păun G (2007) Spiking neural P systems with astrocyte-like control. J Univ Comput Sci 13:1707–1721
Song T, Pan L (2016) Spiking neural P systems with request rules. Neurocomputing 193:193–200
Wang J, Peng H (2013) Adaptive fuzzy spiking neural P systems for fuzzy inference and learning. Int J Comput Math 90:857–868
Zeng X, Zhang X, Pan L (2009) Homogeneous spiking neural P systems. Fundam Inf 97:275–294
Song T, Wang X, Zhang Z, Chen Z (2014) Homogenous spiking neural P systems with anti-spikes. Neural Comput Appl 24(7–8):1833–1841. doi:10.1007/s00521-013-1397-8
Ibarra OH, Păun A, Rodríguez-Patón A (2009) Sequential SNP systems based on min/max spike number. Theor Comput Sci 410:2982–2991
Song T, Xu J, Pan L (2015) On the universality and non-universality of spiking neural P systems with rules on synapses. IEEE Trans NanoBiosci 14:960–966
Song T, Pan L, Păun G (2014) Spiking neural P systems with rules on synapses. Theor Comput Sci 529:82–95
Song T, Pan L (2015) Spiking neural P systems with rules on synapses working in maximum spikes consumption strategy. IEEE Trans NanoBiosci 1:38–44
Song T, Pan L (2015) Spiking neural P systems with rules on synapses working in maximum spiking strategy. IEEE Trans NanoBiosci 4:465–477
Ionescu M, Sburlan D (2007) Several applications of spiking neural P systems. In: Fifth brainstorming week on membrane computing, Sevilla
Adl A, Badr A, Farag I (2010) Towards a spiking neural P systems OS. arXiv preprint arXiv:1012.0326
Zeng X, Song T, Zhang X, Pan L (2012) Performing four basic arithmetic operations with spiking neural P systems. IEEE Trans NanoBiosci 11:366–374
Zhang G, Rong H, Neri F, Pérez-Jiménez MJ (2014) An optimization spiking neural P system for approximately solving combinatorial optimization problems. Int J Neural Syst 24(5):1440006
Wang T, Zhang G, Zhao J, He Z, Wang J, Pérez-Jiménez MJ (2014) Fault diagnosis of electric power systems based on fuzzy reasoning spiking neural P systems. IEEE Trans Power Syst 30:1182–1194
Peng H, Wang J, Pérez-Jiménez MJ, Wang H, Shao J, Wang T (2013) Fuzzy reasoning spiking neural P system for fault diagnosis. Inf Sci 235:106–116
Wang J, Shi P, Peng H, Pérez-Jiménez MJ, Wang T (2013) Weighted fuzzy spiking neural P systems. IEEE Trans Fuzzy Syst 21:209–220
Ishdorj T-O, Leporati A, Pan L, Zeng X, Zhang X (2010) Deterministic solutions to QSAT and Q3SAT by spiking neural P systems with pre-computed resources. Theor Comput Sci 411:2345–2358
Pan L, Păun G, Perez-Jimenez MJ (2011) Spiking neural P systems with neuron division and budding. Sci China Inform Sci 54:1596–1607
Wang X, Song T, Gong F, Zheng P (2016) On the computational power of spiking neural P systems with self-organization. Sci Rep. doi:10.1038/srep27624
Leporati A, Mauri G, Zandron C, Păun G, Pérez-Jiménez MJ (2009) Uniform solutions to SAT and subset sum by spiking neural P systems. Nat Comput 8:681–702
Macias-Ramos LF, Perez-Hurtado I, Garcia-Quismondo M, Valencia-Cabrera L, Perez-Jimenez MJ, Riscos-Nunez A (2012) A P-lingua based simulator for spiking neural P systems. Lect Notes Comput Sci 7184:257–281
Ramirez-Martinez D, Gutierrez-Naranjo MA (2007) A software tool for dealing with spiking neural P systems. In: Gutirrez-Naranjo MA (ed) Proceeding of the 5th brainstorming week on membrane computing, pp 299–313
Macias-Ramos LF, Perez-Jimenez MJ, Song T, Pan L (2015) Extending simulation of asynchronous spiking neural P systems in P-Lingua. Fundam Inform 136:253–267
Tingfang W, Xun W, Zheng Z, Faming G, Tao S, Zhihua C (2016) NES-REBS: a novel nuclear export signal prediction method using regular expressions and biochemical properties. J Bioinform Comput Biol (in press)
Prieto G, Fullaondo A, Rodriguez JA (2014) Prediction of nuclear export signals using weighted regular expressions (wregex). Bioinformatics 30:1220–1227
Carla HV, Chiodi G (2013) Structural characterization of netnes glycopeptide from Trypanosoma cruzi. Carbohydr Res 373:28–34
Păun G, Rozenberg G, Salomaa A (2010) The Oxford handbook of membrane computing. Oxford University Press, Oxford
Gutierrez-Naranjo MA, Perez-Jimenez MJ (2009) Hebbian learning from spiking neural P systems view. Lect Notes Comput Sci 5391:217–230
Zeng X, Zhang X, Zou Q (2016) Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Brief Bioinform 17(2):193–203
Zou Q, Li J, Song L, Zeng X, Wang G (2016) Similarity computation strategies in the microRNA disease network: a survey. Brief Funct Genomics 15(1):55–64
Wang X, Song T, Wang Z, Su Y, Liu X (2013) MRPGA: motif detecting by modified random projection strategy and genetic algorithm. J Comput Theor Nanosci 10:1209–1214
Wang X, Miao Y, Cheng M (2014) Finding motifs in DNA sequences using low-dispersion sequences. J Comput Biol 21:320–329
Zou Q, Hu Q, Guo M, Wang G (2015) HAlign: fast multiple similar DNA/RNA sequence alignment based on the centre star strategy. Bioinformatics 31:2475–2481
Liu B, Chen J, Wang X (2015) Application of learning to rank to protein remote homology detection. Bioinformatics 31:3492–3498
Liu X, Li Z, Liu J, Liu L, Zeng X (2015) Implementation of arithmetic operations with time-free spiking neural P systems. IEEE Trans Nanobioscience 14(6):617–624
Zhang X, Pan L, Paun A (2015) On the universality of axon P systems. IEEE Trans Neural Netw Learn Syst 26:2816–2829
Zhang X, Tian Y, Jin Y (2015) A knee point driven evolutionary algorithm for many-objective optimization. IEEE Trans Evolut Comput 19(6):761–776
Zhang X, Tian Y, Cheng R, Jin Y (2015) An efficient approach to nondominated sorting for evolutionary multiobjective optimization. IEEE Trans Evol Comput 19:201–213
Song T, Pan L (2015) Spiking neural P systems with rules on synapses working in maximum spiking strategy. IEEE Trans NanoBiosci 14:465–477
Zeng X, Zhang X, Song T, Pan L (2014) Spiking neural P systems with thresholds. Neural Comput 26:1340–1361
Gu B, Sheng VS, Tay KY, Romano W, Li S (2015) Incremental support vector learning for ordinal regression. IEEE Trans Neural Netw Learn Syst 26:1403–1415
Gu B, Sun X, Sheng VS (2016) Structural minimax probability machine. IEEE Trans Neural Netw Learn Syst. doi:10.1109/TNNLS.2016.2544779
Wen X, Shao L, Xue Y, Fang W (2015) A rapid learning algorithm for vehicle classification. Inf Sci 295:395–406
Gu B, Sheng VS, Wang Z, Ho D, Osman S, Li S (2015) Incremental learning for support vector regression. Neural Netw 67:140–150
Xia Z, Wang X, Sun X, Wang Q (2016) A secure and dynamic multi-keyword ranked search scheme over encrypted cloud data. IEEE Trans Parallel Distrib Syst 27:340–352
Acknowledgments
This work was supported by National Natural Science Foundation of China (61272152, 61370105, 61402187, 61502535, 61572522 and 61572523), China Postdoctoral Science Foundation funded project (2016M592267), Program for New Century Excellent Talents in University (NCET-13-1031), 863 Program (2015AA020925), and Fundamental Research Funds for the Central Universities (R1607005A).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Z., Zhang, P., Wang, X. et al. A computational approach for nuclear export signals identification using spiking neural P systems. Neural Comput & Applic 29, 695–705 (2018). https://doi.org/10.1007/s00521-016-2489-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-016-2489-z