Abstract
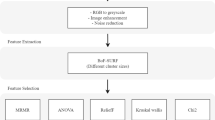
The present paper proposes a novel feature selection technique for the MR brain tumor image classification that aims to choose the optimal feature subset with maximum discriminatory ability in the minimum amount of time. It is based on the fusion of the Fisher and the parameter-free Bat (PFree Bat) optimization algorithm. As the conventional Bat algorithm is bad at exploration, a modification is proposed that guides the Bat by the pulse frequency, global best and the local best position. This improved version of Bat referred to as the PFree Bat algorithm eliminates the velocity equation and directly updates the Bat position. Subsequently, this method in conjunction with the Fisher criteria has been used to select the best set of features for brain tumor classification. The chosen features are then fed to the commonly used least square (LS) support vector machine (SVM) classifier to categorize the area of interest into the high or low grade. For the evaluation of the proposed attribute selection method, tenfold cross-validation has been conducted on a set of 95 ROIs taken from the BRATS 2012 dataset. On an extensive comparison with the other hybrid approaches, the proposed approach brought about the 100% recognition rate in the smallest amount of time. Furthermore, an integrated index is proposed that uniquely identifies the best performing algorithm, taking into account the accuracy, number of features and the computational time. For the fair comparison, the performance of the proposed method has also been examined on breast cancer dataset taken from UCI repository. The obtained results validate that the designed algorithm has better average accuracy than existing state-of-the-art works.





Similar content being viewed by others
References
Zacharaki EI, Wang S, Chawla S, Soo D (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62:1609–1618
Arakeri MP, Reddy GRM (2013) Computer-aided diagnosis system for tissue characterization of brain tumor on magnetic resonance images. Signal Image Video Process 7:1–17
Georgiadis P, Cavouras D, Kalatzis I et al (2009) Enhancing the discrimination accuracy between metastases, gliomas and meningiomas on brain MRI by volumetric textural features and ensemble pattern recognition methods. Magn Reson Imaging 27:120–130
Zhang N, Ruan S, Lebonvallet S et al (2011) Kernel feature selection to fuse multi-spectral MRI images for brain tumor segmentation. Comput Vis Image Underst 115:256–269
Ahmed S, Iftekharuddin KM, Vossough A (2011) Efficacy of texture, shape, and intensity feature fusion for posterior-fossa tumor segmentation in MRI. IEEE Trans Inf Technol Biomed 15:206–213
Sachdeva J, Kumar V, Gupta I et al (2011) Multiclass brain tumor classification using GA-SVM. Dev E-Syst Eng 2011:182–187
Jothi G, Inbarani H (2015) Hybrid Tolerance Rough Set-Firefly based supervised feature selection for MRI brain tumor image classification. Appl Soft Comput J 46:639–651
Liu X, Ma L, Song L et al (2015) Recognizing common CT imaging signs of lung diseases through a new feature selection method based on Fisher criterion and genetic optimization. IEEE J Biomed Heal Inform 19:635–647
Yang X-S (2010) A new metaheuristic bat-inspired algorithm. In: Nature inspired cooperative strategies for optimization (NICSO 2010). Springer, Berlin, pp 65–74
Yilmaz S, Kucuksille EU (2015) A new modification approach on bat algorithm for solving optimization problems. Appl Soft Comput 28:259–275
Bakwad KM, Pattnaik SS, Sohi BS et al (2009) Hybrid bacterial foraging with parameter free PSO. In: Nature and biologically inspired computing 2009. NaBIC 2009. World Congress on, pp 1077–1081
Ramana Murthy G, Senthil Arumugam M, Loo CK (2009) Hybrid particle swarm optimization algorithm with fine tuning operators. Int J Bio-Inspired Comput 1:14–31
Menze BH, Jakab A, Bauer S et al (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024
Lee MC, Nelson SJ (2008) Supervised pattern recognition for the prediction of contrast-enhancement appearance in brain tumors from multivariate magnetic resonance imaging and spectroscopy. Artif Intell Med 43:61–74
Yao X, Liu Y, Lin G (1999) Evolutionary programming made faster. IEEE Trans Evol Comput 3:82–102
Yilmaz S, Kucuksille EU (2013) Improved bat algorithm (IBA) on continuous optimization problems. Lect Notes Softw Eng 1:279–283
Fister Jr I, Fister D, Yang X-S (2013) A hybrid bat algorithm. arXiv Prepr. arXiv1303.6310
Fister Jr I, Fister D, Fister I (2013) Differential evolution strategies with random forest regression in the bat algorithm. In: Proceedings of the 15th annual conference companion on Genetic and evolutionary computation, pp 1703–1706
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC 3:610–621
Vidya KS, Ng EY, Acharya UR et al (2015) Computer-aided diagnosis of myocardial infarction using ultrasound images with DWT, GLCM and HOS methods: a comparative study. Comput Biol Med 62:86–93
Tang X (1998) Texture Information in run-length matrices. IEEE Trans Image Process 7:1602–1609
Amadasun M, King R (1989) Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 19:1264–1274
Tencer L, Reznakova M, Cheriet M (2012) A new framework for online sketch-based image retrieval in web environment. In: Information science, signal processing and their applications (ISSPA), 2012 11th international conference on. IEEE, Montreal, QC, pp 1430–1431
Costa AF, Humpire-mamani G, Juci A et al (2012) An efficient algorithm for fractal analysis of textures. In: 25th SIBGRAPI conference on graphics patterns images. IEEE, Ouro Preto, Brazil, pp 39–46
Hong L, Wan Y, Jain A (1998) Fingerprint image enhancement: algorithm and performance evaluation. IEEE Trans Pattern Anal Mach Intell 20:777–789
Liu Y, Muftah M, Das T et al (2012) Classification of MR tumor images based on Gabor wavelet analysis. J Med Biol Eng 32:22–28
Colominas MA, Schlotthauer G, Torres ME (2014) Improved complete ensemble EMD: a suitable tool for biomedical signal processing. Biomed Signal Process Control 14:19–29
De Brabanter K, Karsmakers P, Ojeda F, et al (2011) LS-SVMlab toolbox user’s guide. ESAT-SISTA technical report 10
Yang X-S, Press L (2010) Nature-inspired metaheuristic algorithms, second edition
Yang X-S, Hossein Gandomi A (2012) Bat algorithm: a novel approach for global engineering optimization. Eng Comput 29:464–483
Meng X-B, Gao XZ, Liu Y, Zhang H (2015) A novel bat algorithm with habitat selection and Doppler effect in echoes for optimization. Expert Syst Appl 42:6350–6364
Karaboga D (2005) An idea based on honey bee swarm for numerical optimization
Karaboga D, Basturk B (2007) A powerful and efficient algorithm for numerical function optimization: artificial bee colony (ABC) algorithm. J Glob Optim 39:459–471
Karaboga D, Basturk B (2008) On the performance of artificial bee colony (ABC) algorithm. Appl Soft Comput 8:687–697
Karaboga D, Akay B (2009) A comparative study of artificial bee colony algorithm. Appl Math Comput 214:108–132
Yang X-S, Deb S (2009) Cuckoo search via Lévy flights. In: Nature & biologically inspired computing 2009. NaBIC 2009. World Congress on, pp 210–214
Yang X-S, Deb S (2010) Engineering optimisation by cuckoo search. Int J Math Model Numer Optim 1:330–343
Mahdavi M, Fesanghary M, Damangir E (2007) An improved harmony search algorithm for solving optimization problems. Appl Math Comput 188:1567–1579
Mangasarian OL, Setiono R, Wolberg WH (1990) Pattern recognition via linear programming: theory and application to medical diagnosis. Large-Scale Numer Optim 22–31
Mangasarian OL, Street WN, Wolberg WH (1995) Breast cancer diagnosis and prognosis via linear programming. Oper Res 43:570–577
Wolberg WH, Street WN, Heisey DM, Mangasarian OL (1995) Computer-derived nuclear features distinguish malignant from benign breast cytology. Hum Pathol 26:792–796
Ratanamahatana CA, Gunopulos D (2003) Feature selection for the naive bayesian classifier using decision trees. Appl Artif Intell 17:475–487
Palaniappan S, Pushparaj T (2013) A novel prediction on breast cancer from the basis of association rules and neural network. Int J Comput Sci Mob Comput 2:269–277
Miao D, Gao C, Zhang N, Zhang Z (2011) Diverse reduct subspaces based co-training for partially labeled data. Int J Approx Reason 52:1103–1117
Abdel-Aal RE (2005) GMDH-based feature ranking and selection for improved classification of medical data. J Biomed Inform 38:456–468
Luukka P, Leppälampi T (2006) Similarity classifier with generalized mean applied to medical data. Comput Biol Med 36:1026–1040
Ahmad F, Isa NAM, Hussain Z, Sulaiman SN (2012) A genetic algorithm-based multi-objective optimization of an artificial neural network classifier for breast cancer diagnosis. Neural Comput Appl 23:1427–1435
Sheikhpour R, Agha M, Sheikhpour R (2016) Particle swarm optimization for bandwidth determination and feature selection of kernel density estimation based classifiers in diagnosis of breast cancer. Appl Soft Comput 40:113–131
Mojtaba S, Bamakan H, Gholami P (2014) A novel feature selection method based on an integrated data envelopment analysis and entropy model. Procedia Comput Sci 31:632–638
Xue B, Zhang M, Browne WN (2012) New fitness functions in binary particle swarm optimisation for feature selection. In: IEEE congress on evolutionary computation (CEC), 2012, pp 1–8
Xue B, Zhang M, Browne WN (2014) Particle swarm optimisation for feature selection in classification: novel initialisation and updating mechanisms. Appl Soft Comput 18:261–276
Maldonado S, Weber R, Basak J (2011) Simultaneous feature selection and classification using kernel-penalized support vector machines. Inf Sci 181:115–128
Miao D, Gao C, Zhang N, Zhang Z (2011) Diverse reduct subspaces based co-training for partially labeled data. Int J Approx Reason 52:1103–1117
Luukka P, Leppalampi T (2006) Similarity classifier with generalized mean applied to medical data. Comput Biol Med 36:1026–1040
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
‘The authors declare that they have no conflict of interest.
Appendix
Appendix
See Table 10.
Rights and permissions
About this article
Cite this article
Kaur, T., Saini, B.S. & Gupta, S. A novel feature selection method for brain tumor MR image classification based on the Fisher criterion and parameter-free Bat optimization. Neural Comput & Applic 29, 193–206 (2018). https://doi.org/10.1007/s00521-017-2869-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-017-2869-z