Abstract
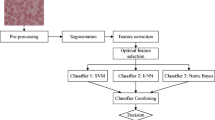
Malaria, being a life-threatening disease caused by parasites, demands its rapid and accurate diagnosis. In this paper, we develop a computer-assisted malaria-infected life-cycle stages classification based on a hybrid classifier using thin blood smear images. The major issues are: feature extraction, feature selection and classification of erythrocytes infected with different life-cycle stages of malaria. Feature set (134 dimensional features) has been defined by the combination of the proposed features along with the existing features. Features such as prediction error, co-occurrence of linear binary pattern, chrominance channel histogram, R–G color channel difference histogram and Gabor features are the newly proposed features in our system. In the feature selection, a two-stage algorithm utilizing the filter method to rank the feature, along with the incremental feature selection technique, has been analyzed. Moreover, the performance of all the individual classifiers (Naive Bayes, support vector machine, k-nearest neighbors and artificial neural network) is evaluated. Finally, the three individual classifiers are combined to develop a hybrid classifier using different classifier combining techniques. From the experimental results, it may be concluded that hybrid classifier formed by the combination of SVM, k-NN and ANN with majority voting technique provides satisfactory results compared to other individual classifiers as well as other hybrid model. An accuracy of 96.54 ± 0.73% has been achieved on the collected clinical database. The results show an improvement in accuracy (11.62, 6.7, 3.39 and 2.39%) as compared to the state-of-the-art individual classifiers, i.e., Naive Bayes, SVM, k-NN and ANN, respectively.











Similar content being viewed by others
References
Cuomo MJ, Noel LB, White DB (2012) Diagnosing medical parasites: a public health officers guide to assisting laboratory and medical officers. http://www.phsource.us/PH/PARA/DiagnosingMedicalParasites
Dhiman S, Baruah I, Singh L (2010) Military malaria in northeast region of India. Defence Sci J 60(2):213–218
Devi SS, Kumar R, Laskar RH (2015) Recent advances on erythrocyte image segmentation for biomedical applications. In: Fourth international conference on soft computing for problem solving, Springer, India pp 353–359
Sertel O, Dogdas Chui BCS, Gurcan MN (2011) Microscopic image analysis for quantitative characterization of muscle fiber type composition. Comput Med Imag Grap 35(7–8):616–628
Ahangi A, Karamnejad M, Mohammadi N et al (2013) Multiple classifier system for EEG signal classification with application to brain–computer interfaces. Neural Comput Appl 23(5):1319–1327
El-Baz AH (2015) Hybrid intelligent system-based rough set and ensemble classifier for breast cancer diagnosis. Neural Comput Appl 26(2):437–446
Di Ruberto C, Dempster A, Khan S, Jarra B (2002) Analysis of infected blood cell images using morphological operators. Image Vision Comput 20(2):133–146
Nicholas RE, Charles JP, David MR, Adriano GD (2006) Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput 44(5):427–436
Devi SS, Sheikh SA, Laskar RH (2016) Erythrocyte features for malaria parasite detection in microscopic images of thin blood smear: a review. Int J Interact Multimed Artif Intel 4(2):35–39
Tek FB, Dempster AG, Kale I (2006) Malaria parasite detection in peripheral blood images. In: British machine vision conference, UK, pp 347–356
Tek FB, Dempster AG, Kale I (2010) Parasite detection and identification for automated thin blood film malaria diagnosis. Comput Vis Image Underst 114(1):21–32
Diaz G, Gonzalez FA, Romero E (2007) Infected cell identification in thin blood images based on color pixel classification: comparison and analysis. In: Iberoamericann congress on pattern recognition, CIARP, 2007, Springer, pp 812–821
Diaz G, Gonzalez FA, Romero E (2009) A semi-automatic method for quantification and classification of erythrocytes infected with malaria parasites in microscopic images. J Biomed Inform 42(2):296–307
Springl V (2009) Automatic malaria diagnosis through microscopic imaging. Faculty of Electrical Engineering, Prague
Khan MI, Acharya B, Singh BK, Soni J (2011) Content based image retrieval approaches for detection of malarial parasite in blood images. Int J Biom Bioinform 5(2):97–110
Soni J, Mishra N, Kamargaonkar NC (2011) Automatic difference between RBC and malaria parasites based on morphology with first order features using image processing. Int J Adv Eng Tech 1(5):290–297
Prasad K, Winter J, Bhat UM, Acharya RV, Prabhu GKJ (2012) Image analysis approach for development of a decision support system for detection of malaria parasites in thin blood smear images. J Digit Imaging 25(4):542–549
Abdul-Nasir AS, Mashor MY, Mohamed Z (2013) Colour image segmentation approaches for detection of malaria parasites using various colour models and k-means clustering. WSEAS T Biol Biomed 10:41–55
Savkare S, Narote S (2011) Automatic detection of malaria parasites for estimating parasitemia. Int J Comput Sci Secur 5(3):310–315
Das DK, Ghosh M, Pal M, Maiti AK, Chakraborty C (2013) Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron 45:97–106
Ghosh M, Das DK, Chakraborty C, Ray AK (2013) Quantitative characterisation of Plasmodium vivax in infected erythrocytes: a textural approach. Int J Artif Intell Soft Co 3:203–221
Das DK, Maiti AK, Chakraborty C (2012) Textural pattern classification of microscopic images for malaria screening. Advances in Therapeutic Engineering. CRC Press, Boca Raton, pp 419–446
Maity M, Maiti AK, Dutta PK, Chakraborty C (2012) A web accessible framework for automated storage with compression and textural classification of malaria parasite images. Int J Comput Appl 52(15):31–39
Kumarasamy SK, Ong SH, Tan KSW (2011) Robust contour reconstruction of red blood cells and parasites in the automated identification of the stages of malarial infection. Mach Vision Appl 22:461–469
Das DK, Ghosh M, Chakraborty C, Maiti AK, Pal M (2011) Probabilistic prediction of malaria using morphological and textural information. In: International conference on image information processing (ICIIP), IEEE India
Memeu DM (2014) A rapid malaria diagnostic method based on automatic detection and classification of plasmodium parasites in stained thin blood smear images. Dissertation, University of Nairobi
Gitonga L, Memeu DM, Kaduki KA, Kale MAC, Muriuki NS (2014) Determination of plasmodium parasite life stages and species in images of thin blood smears using artificial neural network. Open J Clin Diag 4:78–88
Purwar Y, Shah SL, Clarke G, Almugairi A, Muehlenbachs A (2011) Automated and unsupervised detection of malarial parasites in microscopic images. Malaria J 10:364
Annaldas S, Shirgan SS, Marathe VR (2014) Automatic identification of malaria parasites using image processing. Int J Emerg Eng Res Technol 2:107–112
Bairagi VK, Charpe KC (2016) Comparison of texture features used for classification of life stages of malaria parasite. Int J Biomed Imaging. Article ID 7214156
Central for diseases control and prevention (2013). USA. http://www.cdc.gov/dpdx/malaria/gallery.html. Accessed 21 May 2015
Somasekar J, Reddy BE (2015) Segmentation of erythrocytes infected with malaria parasites for the diagnosis using microscopy imaging. Comput Electr Eng 45:336–351
Thangavel K, Manavalan R, Aroquiaraj IL (2009) Removal of speckle noise from ultrasound medical image based on special filters: comparative study. ICGST-GVIP J 9(3):25–32
Devi SS, Roy A, Singha J, Sheikh SA, Laskar RH (2016) Malaria infected erythrocyte classification based on a hybrid classifier using microscopic images of thin blood smear. Multimed Tools Appl. doi:10.1007/s11042-016-4264-7
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Sys Man Cyber 9(1):62–66
Jung C, Kim C (2010) Segmenting clustered nuclei using h-minima transform- based marker extraction and contour parameterization. IEEE Trans Biomed Eng 57(10):2600–2604
Roy A, Singha J, Devi SS, Laskar RH (2016) Impulse noise removal using SVM classification based fuzzy filter from gray scale images. Signal Process 128:262–273
Proakis JG, Manolakis DG (2001) Digital signal processing: principles algorithms and applications. Prentice-Hall, Upper Saddle River
Chowdhury S, Verma B, Stockwell D (2015) A novel texture feature based multiple classifier technique for roadside vegetation classification. Expert Syst Appl 42(12):5047–5055
Hahnel M, Klunder D, Kraiss, Color K-F (2004) Texture features for person recognition. In: IEEE international joint conference on neural networks 2004, pp 647–652
Terrillon JC, Shirazi MN, Fukamachi H, Akamatsu S (2000) Comparative performance of different skin chrominance models and chrominance spaces for the automatic detection of human faces in color images. In: 4th IEEE automatic face and gesture recognition, 2000, pp 54–61
Siggelkow S (2002) Feature histograms for content-based image retrieval. Dissertation, Universitat Freiburg
Bashyal S, Venayagamoorthy GK (2008) Recognition of facial expressions using Gabor wavelets and learning vector quantization. Eng Appl Artif Intell 21(7):1056–1064
Theodoridis S, Pikrakis A, Koutroumbas K, Cavouras D (2010) Introduction to pattern recognition: a matlab approach, 4th edn. Academic Press, New York, pp 107–135
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40(1):16–28
Ding H, Feng PM, Chen W, Lin H (2014) Identification of bacteriophage virion proteins by the ANOVA feature selection and analysis. Mol BioSyst 10(8):2229–2235
Niu B, Huang G, Zheng L, Wang X, Chen F, Zhang Y, Huang T (2012) Prediction of substrate-enzyme-product interaction based on molecular descriptors and physicochemical properties. J Proteomics 75:1654–1665
Settouti N, Bechar MEA, Chikh MA (2016) Statistical comparisons of the top 10 algorithms in data mining for classification task. Int J Interactive Multimed Artif Intel 4(1):46–51
Kumari P, Vaish A (2016) Feature-level fusion of mental task’s brain signal for an efficient identification system. Neural Comput Appl 27(3):659–669
Russell S, Norvig P (2003) Artificial intelligence: a modern approach, 2nd edn. Prentice Hall, Englewood Cliffs. ISBN 978-0137903955
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Burges CJC (1998) A tutorial on support vector machines for pattern recognition. Data Mining Knowl Discov 2:121–167
Ahmad I, Hussain M, Alghamdi A et al (2014) Enhancing SVM performance in intrusion detection using optimal feature subset selection based on genetic principal components. Neural Comput Appl 24(7):1671–1682
Pujari JD, Yakkundimath R, Byadgi AS (2016) SVM and ANN based classification of plant diseases using feature reduction technique. Int J Interact Multimed Artif Intel 3(7):6–14
Altman NS (1992) An introduction to kernel and nearest-neighbor non parametric regression. Am Stat 46(3):175–185
Weinberger KQ, Saul LK (2009) Distance metric learning for large margin nearest neighbor classification. J Mach Learn Res 10:207–244
Omranpour H, Ghidary SS (2016) A heuristic supervised euclidean data difference dimension reduction for KNN classifier and its application to visual place classification. Neural Comput Appl 27(7):1867–1881
Duda RO, Hart PE, Stork DG (2001) Pattern classification. Wiley, New Delhi
Semwal VB, Mondal K, Nandi GC (2015) Robust and accurate feature selection for humanoid push recovery and classification: deep learning approach. Neural Comput Appl. doi:10.1007/s00521-015-2089-3
Semwal VB, Raj M, Nandi GC (2015) Biometric gait identification based on a multilayer perceptron. Robot Auton Syst 65:65–75
Gomez AB, Mingueza NL, García del Pozo MC (2015) OpinAIS: an artificial immune system-based framework for opinion mining. Int J Interact Multimed Artif Intel 3(3):25–34
Kuncheva LI (2004) Combining pattern classifiers: methods and algorithms, 2nd edn. Wiley, New Jersey
Ruta D, Gabrys B (2005) Classifier selection for majority voting. Inform Fusion 6(1):63–81
Singha J, Laskar RH (2015) Self co-articulation detection and trajectory guided recognition for dynamic hand gestures. IET Comput Vision 10(2):143–152
Singha J, Laskar RH (2016) Hand gesture recognition using two-level speed normalization, feature selection and classifier fusion. Multimedia Syst. doi:10.1007/s00530-016-0510-0
Singha J, Laskar RH (2016) Recognition of global hand gestures using self co-articulation information and classifier fusion. J Multimodal User Interfaces 10(1):77–93
Singha J, Roy A, Laskar RH (2016) Dynamic hand gesture recognition using vision-based approach for human–computer interaction. Neural Comput Appl. doi:10.1007/s00521-016-2525-z
Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH (2007) A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg 77(6):119–127
Murphy SC, Shott JP, Parikh S, Etter P, Prescott WR, Stewart VA (2013) Review article: malaria diagnostics in clinical trials. Am J Trop Med Hyg 89(5):824–839
Acknowledgements
This work is supported by the Speech and Image Processing Lab under Department of ECE at National Institute of Technology, Silchar, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest with any organization/institute. The authors do not have any secondary interest from the materials discussed in this manuscript.
Rights and permissions
About this article
Cite this article
Devi, S.S., Laskar, R.H. & Sheikh, S.A. Hybrid classifier based life cycle stages analysis for malaria-infected erythrocyte using thin blood smear images. Neural Comput & Applic 29, 217–235 (2018). https://doi.org/10.1007/s00521-017-2937-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-017-2937-4